Science
109 The Suppression of Signal-Switching Defects in the Escherichia Coli Serine Chemoreceptor
Ana Rowe and John Parkinson (School of Biological Sciences)
Faculty Mentor: John Parkinson (School of Biological Sciences, University of Utah)
ABSTRACT
The model bacterium Escherichia coli contains a chemotaxis system that allows the cell to change its swimming behavior in response to its environment. As the cell swims, transmembrane chemoreceptors detect concentration changes in attractant and repellent compounds and transmit signals across the inner membrane that elicit an appropriate locomotor response. Previous work identified an amino acid near the cytoplasmic tip of the receptor (F396) that plays a critical role in enabling Tsr to modulate its signal output in response to a serine stimulus. The objective of my project is to characterize second-site mutations that rescue the chemotactic ability of F396 receptor mutants. These suppressors presumably enable mutant receptors to undergo the conformational changes necessary for serine sensing and signaling. To identify such suppressors, I subjected plasmids that encode Tsr-F396G (glycine) or Tsr-F396W (tryptophan) mutant receptors to random mutagenesis and selected for mutant plasmids that promoted improved serine chemotaxis. DNA sequence analysis of the revertant plasmids showed that most of the suppressor mutations lie near the hairpin tip of Tsr. I obtained few F396W revertants and none with substantially improved function in contrast to F396G, which gave rise to more revertants with better chemotactic ability. In subsequent experiments, I determined the expression levels of the doubly mutant Tsr proteins and characterized their signaling properties with in vitro FRET-based kinase assays. My findings served to identify Tsr structural features important for signal control of the cytoplasmic tip and enabled me to develop a more complete model of transmembrane signaling by the Tsr protein.
INTRODUCTION
In the microbial world, as in all of life, organisms detect and process information about their environment. To internalize meaningful messages of external stimuli, microorganisms require extraordinarily sensitive sensory systems. Such systems enable motile microbes to move toward favorable chemical environments and away from noxious chemicals, a behavior known as chemotaxis. Specialized transmembrane receptors facilitate sensing and responding to these environmental cues. These receptors, called methyl-accepting chemotaxis proteins (MCPs), initiate a communication cascade of signaling steps that ultimately control the cell’s locomotion.
Escherichia coli is a motile bacterium that swims by rotating its flagellar filaments. The rotary motors can turn in both clockwise (CW) and counterclockwise (CCW) directions (Figure 1A). CCW rotation enables the flagellar propellers to form a bundle that pushes the cell forward in a “run”. A CW reversal of one or more motors causes the bundle to disperse and the cells to execute random turning motions, termed a “tumble”. When swimming in the absence of a chemical gradient, the flagellar motors switch unpredictably between CW and CCW rotation, causing the cell to move about in a random-walk fashion. In an attractant gradient whenever the cell heads up-gradient, it senses that attractant concentration is increasing over time and this information produces a signal that lowers the probability of the next tumbling event. Thus, E. coli cells move up-gradient in a biased random walk fashion (Figure 1B). E. coli has five MCP-type chemoreceptors that sense different chemicals. The serine chemoreceptor, Tsr, and the other receptors function as membrane-spanning homodimers with a cytoplasmic signaling domain (Figure 2; Parkinson et al, 2015). Three receptor dimers assemble trimers of dimers through association of their cytoplasmic tips. The trimer tips provide an interaction surface for an adaptor protein, CheW, that in turn couples an autokinase, CheA, to chemoreceptor control (Parkinson et al, 2015).
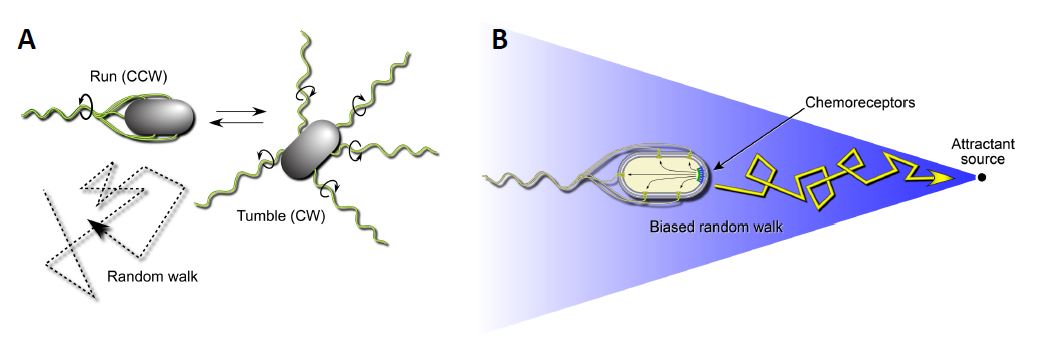
Figure 1: Swimming behaviors of E.coli. A. Transmembrane chemoreceptors detect changes in their surrounding environment and transmit signals that control the cell’s flagellar motors. In the absence of a chemical gradient (right) the cell will swim in a tumbling motion to seek out nutrients. B: Transmembrane chemoreceptors detect changes in their surrounding environment and transmit signals that control the cell’s flagellar motors. In the presence of an attractant source, bacteria such as E. coli can modulate their swimming behavior to favor a forward swimming motion.
Receptor signaling complexes have kinase-ON and kinase-OFF activity states (Figure 3).
When no attractant is bound to the receptors, CheA autophosphorylates, using ATP as the phosphodonor. In this kinase-ON output state, CheA transfers its phosphoryl groups to CheY molecules, which bind to the flagellar motors to promote CW rotation (Figure 3). Phospho-CheY has a short half-life due to a phosphatase partner CheZ, which enables the cell to rapidly change its swimming behavior upon detecting a change in chemoattractant level (Silversmith et al, 2003). Attractant-bound receptors undergo a conformational change that propagates to the associated CheA and stops its autokinase activity (Parkinson et al, 2015). In this kinase-OFF state, phospho- CheY levels quickly plummet, through CheZ activity, and the flagellar motors adopt their default CCW direction of rotation.
Because of their small size, collisions with water molecules (Brownian motion) can knock E. coli cells off-course, so they can only traverse attractant gradients in a biased random-walk fashion. To effectively explore their environment for nutrients, the cells must make temporal comparisons of attractant concentration as they swim about. Reversible covalent modifications of the receptor molecules provide a short-term “memory” of the recent chemical past. Enzymes CheB and CheR catalyze those modifications to adjust receptor output to prevailing chemical conditions (Figure 4). CheR, a methyltransferase, interacts with kinase-OFF receptors and converts specific glutamyl residues to glutamyl methyl esters, which shifts signal output toward the kinase-ON state (Lai et al, 2017). Conversely, the CheB interacts with receptors in the kinase-ON state and shifts them toward kinase-OFF output by hydrolyzing the methylated glutamyl residues (Parkinson et al, 2015). As such, this system acts as a “memory” for the cell so that it can continue to move up the chemical gradient.
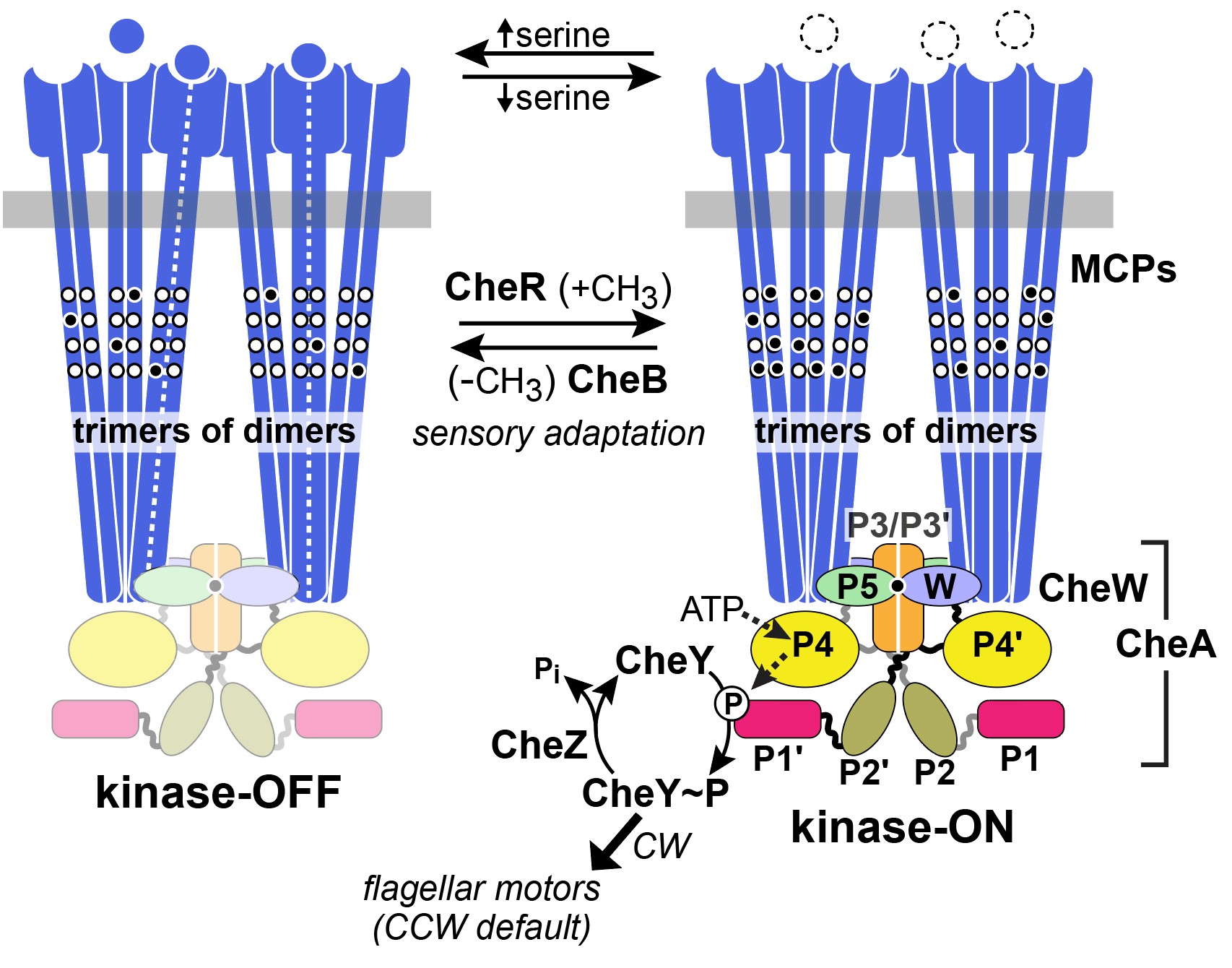
Figure 2: Two-state model of receptor signaling and the chemotaxis phosphorelay pathway. In the absence of a stimulus (right), CheA autophosphorylates and the reaction promotes signaling states that enhance CW flagellar rotation. When an attractant is bound, CheA is dephosphorylated to promote signaling states that augment CCW flagellar rotation. Binding of a ligand or remocing methyl groups shifts the receptor from the kinase-on to the kinase-off state. Attractant release and addition of methyl groups shift the receptor from kinase-off to kinase-on (Parkinson et al, 2015).
MCPs cluster at the cell poles to form large cooperative receptor signaling arrays (Pinas et al, 2022). Array formation enhances chemoreceptor sensitivity by enabling receptors to share and cooperatively act on stimulus information. Signaling interactions between arrayed receptors produce highly cooperative responses to attractant stimuli such as serine (Briegel et al, 2014). Additionally, homodimers of MCPs such as Tsr can assemble into mixed trimers of dimers with other chemoreceptors such as Tar (aspartate and maltose receptor), which further enhances the sensing capabilities of the cell to modulate kinase activity (Ames et al, 2002).
Tsr dimers have an extended four-helix coiled-coil signaling domain with a highly conserved hairpin tip (Figure 3). The tip promotes trimer formation and regulates CheA activity and is thought to play an important role in conformational transitions between the kinase-ON and kinase-OFF output states (Parkinson et al, 2015). The he mechanisms of signal transmission and kinase control by chemorecptors are largely unknown, but a phenylalanine (F) residue, at position 396 in Tsr plays a crucial role. This amino acid is the only universally conserved residue in MCP receptors (Ortega et al, 2013). An X-ray structure of Tsr showed that F396 residues interact across the subunit interface of the dimer tip (Figure 4; Kim et al, 1999). A subsequent all-atom molecular dynamics simulation of the Tsr dimer tip revealed that the F396/F396′ stacking interaction occasionally flipped and that the preferred stacking arrangement in a timer correlated with the receptor’s adaptational modification state. An unmethylated Tsr model favored one arrangement; a highly methylated Tsr model favored the alternative stacking arrangement. Wild- type Tsr flipped between those two states, spending roughly equal time in each. These results suggested that the orientation of F396/F396′ stacking determines the Tsr signaling state and that a flip in the stacking interaction triggers a shift in Tsr output.
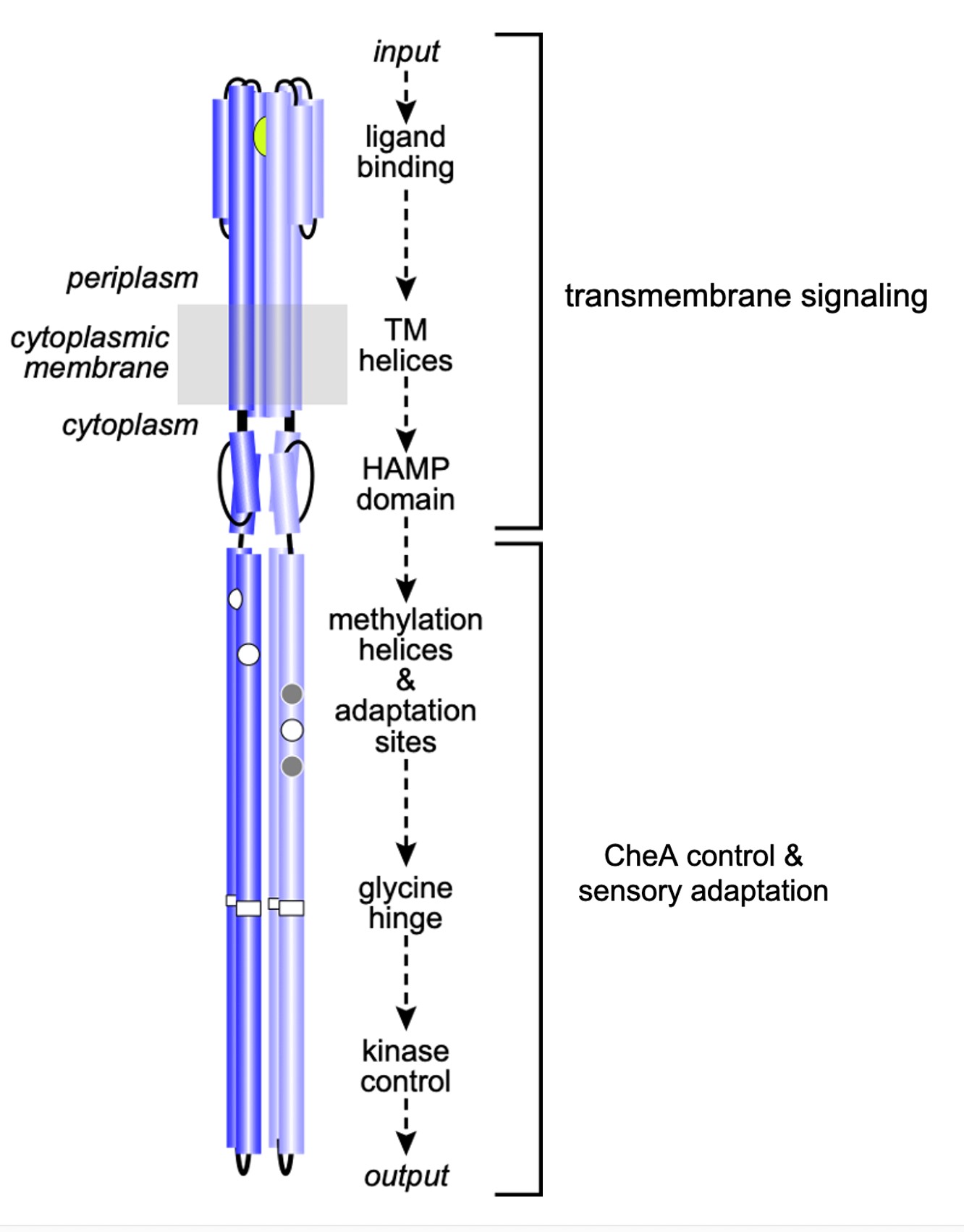
Figure 3: Signaling elements in the Tsr homodimer. Sensory information propagates from the ligand-binding domain to the kinase-control tip through structural interactions between relay elements.
To determine the importance of F396 to Tsr signaling, a series of Tsr mutants with all possible amino acid changes at residue 396 were constructed and assessed for chemotactic ability (Ortega et al, 2013). None of the mutant receptors could support serine chemotaxis, indicating that phenylalanine at this position is indeed critical for proper receptor function. Subsequent studies of Tsr-F396* mutant receptors showed that many of them were strongly shifted toward a kinase-ON output state, suggesting that aromatic stacking of F396/F396′ might play an important role in stabilizing the kinase-OFF signaling state.
Interestingly, glycine (G), the smallest amino acid, and tryptophan (W), the largest, produced similar signaling defects when replacing the wild-type Tsr-F396 residue. Both the Tsr-
F396G and Tsr-F396W receptors were effectively locked in a kinase-ON output state in a cell lacking the sensory adaptation enzymes, CheR and CheB. But in an adaptation-competent host, both mutant receptors were able to shift to kinase-OFF output in response to a large serine stimulus. These results suggest that both the G and W side chains at Tsr residue 396 can support kinase-OFF signaling, but that output conformation may be much less stable than in the wild-type receptor. Conceivably, the inability of the mutant receptors to support chemotaxis, even in a cell with the adaption enzymes, is because they are too strongly biased toward the kinase-ON output state.
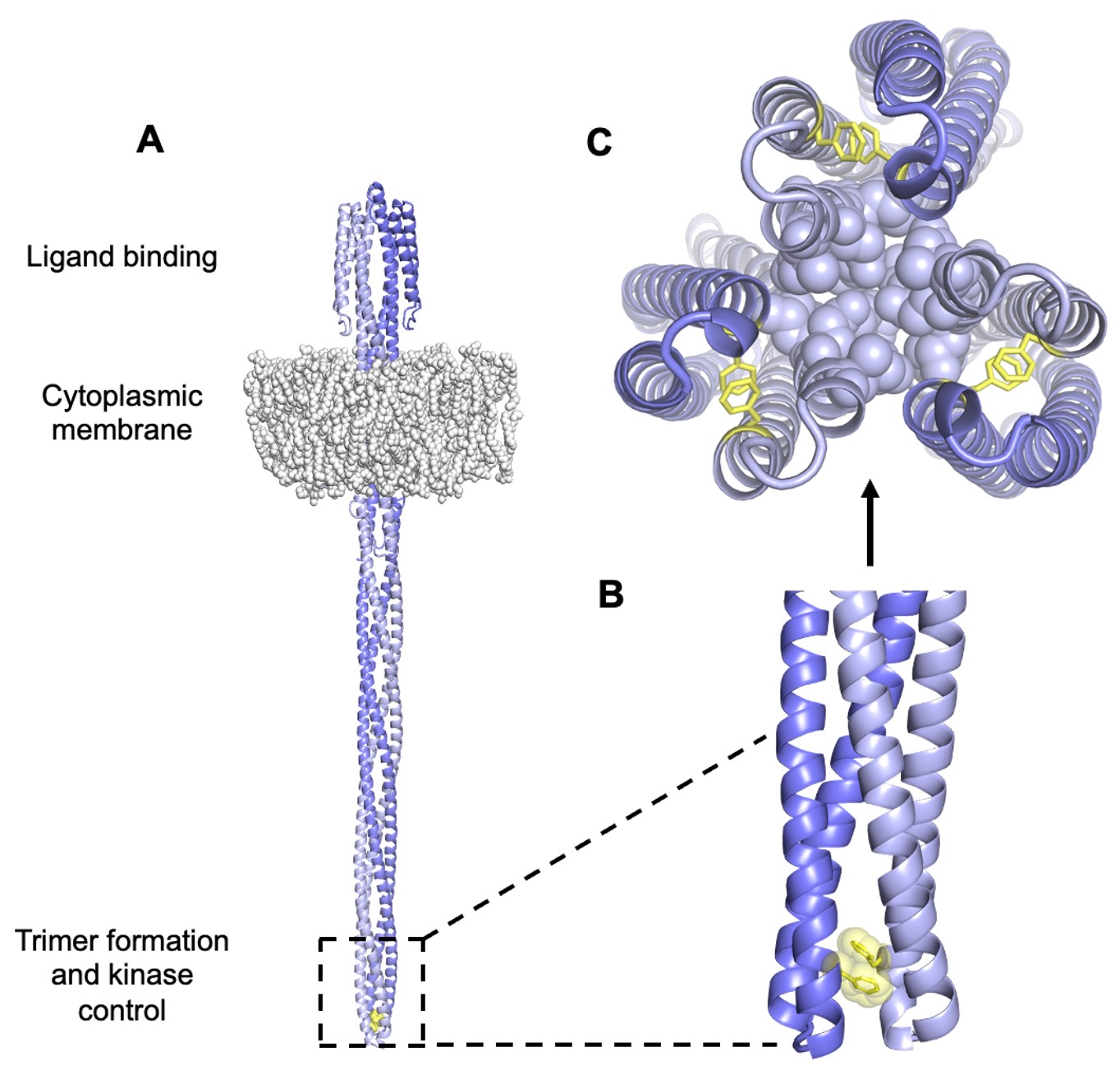
Figure 4: A. Model of Tsr. The hairpin tip circled in black holds amino acid residues that interact with other subunits to form trimer-of-dimers. Encompassed within this region is the conserved amino acid F396. B. These F396 residues lie at the interface between subunits of a dimer to interact in a specific conformation via stacking of its aromatic side chain. C. Tsr residues at the tip interact in trimer-of-dimers to stabilize the tip during kinase-on and kinase-off states.
My research employed reversion analysis of the Tsr-F396G and F396W receptors. I isolated and characterized a number of second-site mutations in the mutant tsr genes that could, to some extent, suppress their defective signaling properties. The signaling properties of the pseudorevertant (double mutant) receptors and of receptors carrying only the suppressor change provided new mechanistic insights into the nature of hairpin tip signaling states and their control by adjacent signaling elements in the Tsr molecule. Moreover, this work has provided a new, testable model of how a chemoreceptor responds to sensory signals.
MATERIALS AND METHODS
Bacterial Strains
E. coli strains used were isogenic derivatives of K-12 RP437 (Parkinson et al, 1982). The relevant genotype of each strain used in subsequent experiments was: UU1623 [Δtap Δtsr Δtrg Δaer]; UU1935 [mutD5]; UU2377 [tsr-R69E Δ(tar-tap) Δtrg Δaer]; UU2378 [tsr-T156K Δ(tar- tap) Δtrg Δaer]; UU2567 [Δ(tar–cheZ) Δtsr Δaer Δtrg]; UU2610 [Δaer ∆(tar-cheB) Δtsr Δtrg]; UU2612 [Δaer Δ(tar-tap) Δtsr Δtrg]; and UU3333 [Δ(tar-tap), Δtsr Δtrg Δaer] (Zhou et al, 2011; Lai et al, 2014).
Plasmids
Plasmids used in this project were: pKG116 (Gosink et al, 2006), which confers chloramphenicol resistance and has a salicylate-inducible expression/cloning site; pPA114, a pKG116 derivative that carries the wild-type tsr gene under salicylate-inducible control (Ames et al, 2002); pVS88, a plasmid compatible with pPA114 derivatives that confers ampicillin resistance and expresses the CheY-YFP and CheZ-CFP proteins under inducible IPTG control (Chang et al, 1978; Sourjik et al, 2007). IPTG is an analog of allolactose to act as an inducer for gene expression in the lac operon (Briand et al, 2016).
Growth media and buffers.
Cultures were grown in L broth (1% tryptone, 0.5% NaCl and 0.5% yeast extract) and colonies were harvested on L plates that contained L broths and 1.5% agar. Tryptone broth contained the same ingredients as L-broths but lacked the yeast extract. Solutions used in in vivo FRET based kinase assays included KEP and Tethering buffer. KEP buffer contained 10 mM KPO4 (pH 7.0), and 0.1 mM K-EDTA. Tethering buffer was comprised of KEP with 10 mM Na- Lactate, 75 mM NaCl, 0.1 mM L-Methionine, and 100 𝜇g/ml chloramphenicol (Parkinson et al,
1982).
Chemotaxis Assays
Serine chemotaxis ability of mutant strains was assessed on semi-solid tryptone agar plates that contained 12.5 µg/ml chloramphenicol and 0.6 µM sodium salicylate (Parkinson, 1980). These plates permit the bacteria to swim in a semi-liquid media. Plates were incubated at 32.5°C for 6 to 8 hours. Mutants were tested for temperature-dependent defects at 25°C and 37°C.
Isolation of chemotactic pseudorevertants
Mutant tsr genes were manipulated and expressed in derivatives of pPA114. Tsr-F396W and Tsr-F396G) plasmids were subjected to random mutagenesis by replication in UU1935.
Plasmid pools from UU1935 were introduced UU2612, and 50 µl aliquots of the transformation mix were streaked across T-swim plates to reveal chemotactic clones after incubation at 32.5°C for 16 hours (Ames et al, 2002). Plasmids were isolated from revertants using QIAGEN DNA extraction kits. Their mutant tsr genes were sequenced by the Protein-DNA Core Facility of the University of Utah.
Expression levels of Mutant Tsr Proteins
Strain UU2610 (CheR-, CheB-) was used as a host to assess Tsr protein expression levels from mutant plasmid derivatives. Cells were grown in tryptone broth with appropriate antibiotics to the mid-exponential phase. Cells were washed, resuspended in Laemmli sample buffer, and lysed by boiling for five minutes (Laemmli, 1970). Samples were analyzed by electrophoresis in denaturing polyacrylamide gels, and Tsr bands were visualized by immunoblotting with a polyclonal rabbit antiserum directed against the Tsr cytoplasmic domain (Mowery et al, 2002; Ames et al, 1994). Tsr bands were quantified using ImageJ software.
Dominance and Jamming Assays
Dominance assays of suppressor mutants were performed in strains UU2377 and UU2378 as described by Ames et al. Jamming assays of suppressor mutants were performed in strain UU1623 as described by Ames et al.
In vivo FRET-based Kinase Assays
The protocol and data analysis for in vivo FRET-based kinase assays followed the methods described by (Sourjik et al, 2007). Mutant Tsr plasmids and the FRET reporter plasmid, pVS88were introduced into strain UU2567. The transformant cells were grown at 30°C for five hours to mid-exponential phase in tryptone broth (50 µg/ml ampicillin, 12.5 µg/ml chloramphenicol, 100 µg/ml sodium salicylate, IPTG). Cells, were washed by centrifugation, bound to a polylysine-treated cover slip, and mounted in a flow cell maintained at 30°C throughout each experiment). Cells were subjected to a series of serine concentrations and the dose-response data were analyzed with Kaleidagraph software. In the absence of a serine response, receptor-generated kinase activity was measured by the FRET change elicited by 3 mM KCN (Lai & Parkinson, 2014).
RESULTS
Pseudoreversion analysis of Tsr-F396G and Tsr-F396W mutant chemoreceptors
To determine whether it is possible to “heal” the functional defects of the Tsr-F396G and Tsr-F396W receptors, I looked for second-site mutations within the mutant tsr coding sequence that could suppress their defects (Figure 5). Mutant receptor genes carried in plasmid pPA114 derivatives were mutagenized in an error-prone mutD host. Treated plasmids were transformed into recptorless host UU2612 and chemotactic pseudorevertants were picked from soft agar plates. These chemotactic pseudorevertants presumably contained suppressor mutations that improved the function of the parental mutant receptor (Figure 5). I then identified the suppressor mutations in the pseudorevertant plasmids by DNA sequence analysis in the tsr coding region.
To isolate the suppressor mutants from the double mutants, I converted the Tsr-F396G and Tsr- F396W mutation in the pseudorevertants back to wild-type (F396) using sequence-targeted PCR. I obtained 3 different suppressor mutations for F396W, and 16 different suppressors for F396G (Figure 6A). All of these mutations were single base pair substitutions. With one exception (D481N), their inferred amino acid replacements fell within the hairpin tip or flexible bundle of Tsr. Second-site suppressors of F369G produced more robust chemotaxis than those of F396W, with several F369G suppressors performing as well as wild-type Tsr on soft agar swim plates (See Appendix; Table 1).
Among Tsr-F396G pseudorevertants, the S327N suppressor was found four times; A323V, D363G, and I455V were each found twice. For Tsr-F396W, A382V and A382T were each obtained twice. Overall, I found an average of about 1.4 mutations per suppressor site, which, if all mutations arose independently, indicates that the mutant hunt was not fully saturated and that there may be additional suppressors yet undiscovered.
Chemotaxis promoted by the mutant chemoreceptors
I tested the serine chemotaxis phenotypes of the doubly-mutant pseudorevertant receptors in soft agar plates (see example in Figure 6B). In the UU2612 host, all F396G pseudorevertant receptors produced a phenotype similar to wild-type Tsr. The double mutant Tsr- F396G/V466T supported chemotaxis better than the wild-type Tsr. The F396W pseudorevertant receptors exhibited only partial chemotactic function. The Tsr-F396W defect is evidently difficult or impossible to fully suppress, in contrast to that of Tsr-F396G.
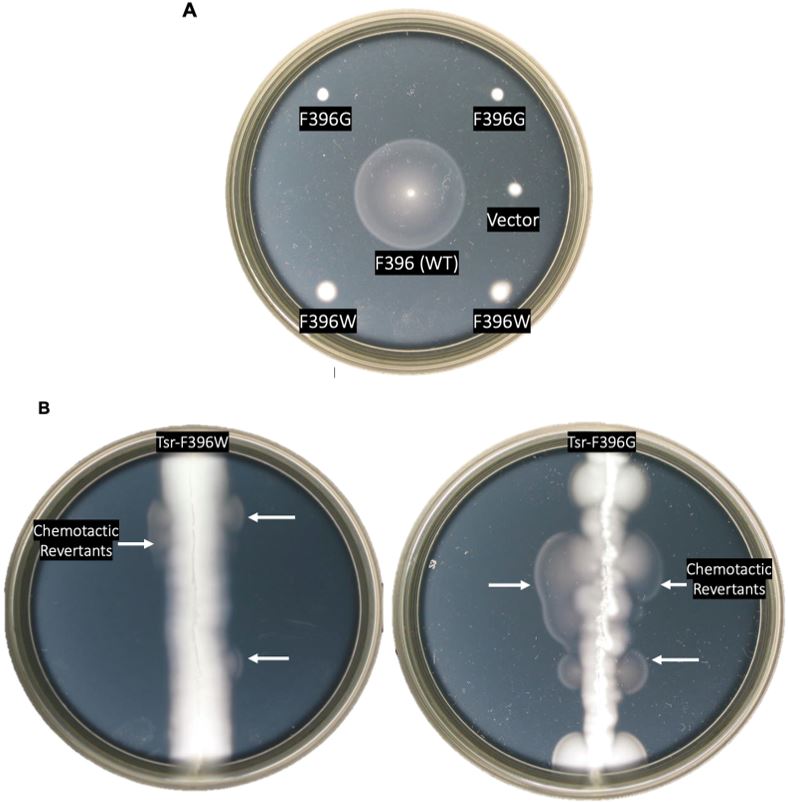
Figure 5: Chemotaxis assays of mutant receptors. A. Chemotactic behaviors of Tsr- F396G and Tsr-F396W mutants as compared to F396 wild-type on soft agar plates. The strains that contained wild-type, mutant, or vector plasmids were all screened for chemotaxis. B. Screening for serine chemotaxis after random mutagenesis in a host that
is Δmcp. White arrows represent swarms containing chemotactic pseudorevertants.
Chemotaxis promoted by the mutant chemoreceptors
I tested the serine chemotaxis phenotypes of the doubly-mutant pseudorevertant receptors in soft agar plates (see example in Figure 6B). In the UU2612 host, all F396G pseudorevertant receptors produced a phenotype similar to wild-type Tsr. The double mutant Tsr- F396G/V466T supported chemotaxis better than the wild-type Tsr. The F396W pseudorevertant receptors exhibited only partial chemotactic function. The Tsr-F396W defect is evidently difficult or impossible to fully suppress, in contrast to that of Tsr-F396G.
To characterize the functional properties of receptors carrying only a suppressor change, I converted the F396 mutant codon in each pseduorevertant back to wild type (F396) using sequence-targeted PCR. In host strain UU2612, 9 out of 17 of the single-mutant receptors exhibited wild-type Tsr function on soft agar plates, 4 out of 17 had partial function, and 4/17 were nonfunctional (See Appendix). None of the suppressors displayed chemotactic function greater than the double-mutants, i.e. F396* plus the second-site mutation. Additional functional tests to determine whether these mutant receptors are capable of making stable protein and characterizing their signaling properties with FRET kinase assays will reveal structural features important for receptor signaling.
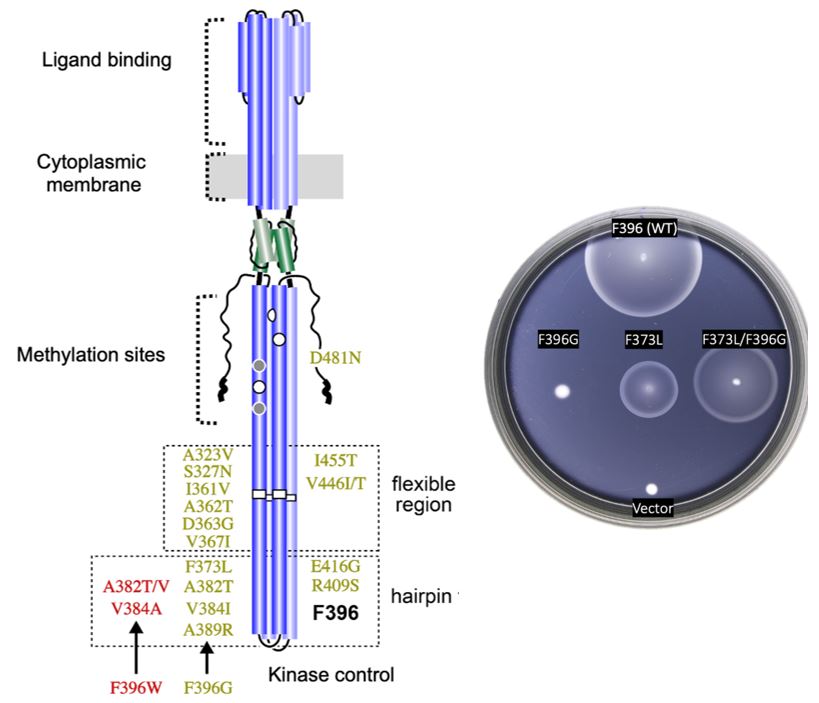
Figure 6: Found suppressor mutants in tsr. A. Summary of the glycine (green) and tryptophan (red) suppressors found within the Tsr receptor. B. Example of chemotactic behavior of the original mutant F396G (left), the isolated suppressor mutant (center), and the double mutant
(right).
Immunoblot Analysis of Mutant Receptors
I measured the intracellular levels of the pseudorevertant and suppressor proteins by gel electrophoresis and anti-Tsr immunoblotting of cell extracts. All mutant proteins had steady-state levels comparable to wild-type Tsr (Figure 8). This result is particularly important for the receptors that failed to support chemotaxis because it means that the mutant protein is expressed normally but cannot function normally.
Signaling Properties of Mutant Tsr Proteins
To elucidate the functional properties of my mutant receptors, I used a FRET-based kinase assay {Sourjik et al, 2002) to determine the ability of the mutant receptors to modulate CheA activity in response to serine stimuli. I expressed the mutant receptors in UU2567, a host that lacks the sensory adaptation enzymes CheR and CheB. In this reporter strain, wild-type Tsr molecule produces relatively high kinase activity that responds to serine stimuli with half- maximal inhibition (K1/2) at ~18 µM (Figure 9A). The signaling behaviors of the mutant receptors were interpreted in the context of a two-state model of kinase-OFF and kinase-ON receptor output states (see Figure 2). Off-shifted mutant receptors have higher serine response sensitivity (lower
K1/2), whereas ON-shifted mutant receptors reduce serine response sensitivity and show higher
K1/2) values or fail to respond at even very high serine concentrations.
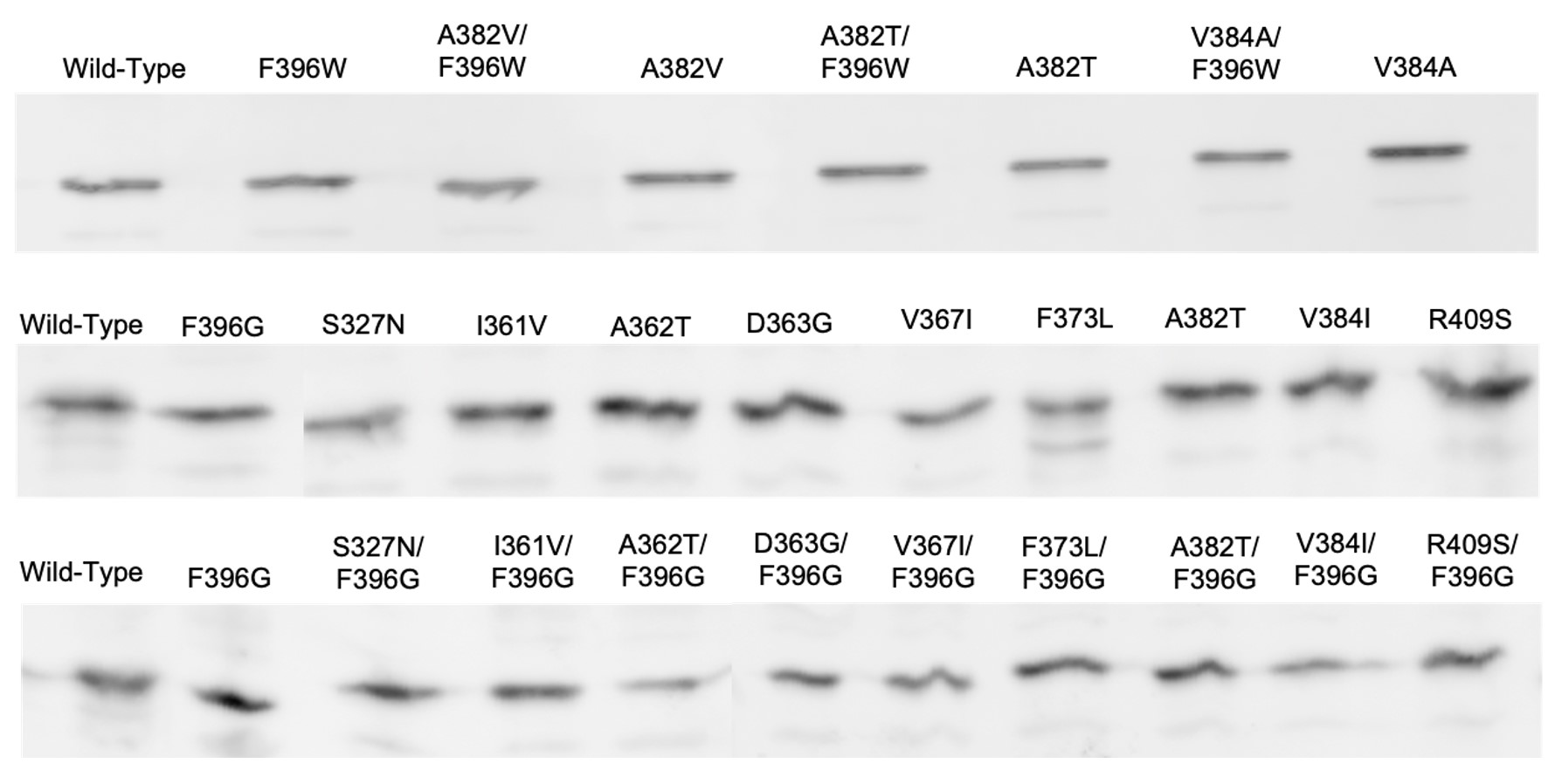
Figure 8: Immunoblots of Tsr-F396G/W double mutants and suppressors. Protein expression samples were expressed in strain UU2610, a host which lacks sensory adaptation enzymes
CheR and CheB. Wild-type Tsr is expressed in the far-left column.
Tsr-F396G exhibited no response to serine in the UU2567 host but did evince some kinase activity when treated with KCN (Figure 9B). This behavior indicates that F396G has a non-responsive kinase-ON output state. Similarly, Tsr-F396W failed to respond to any level of serine concentration but produced kinase-ON output (Figure 9B).
Note that both Tsr-F396* mutant receptors only produced about 50% of the Tsr wild-type kinase activity. This is an important clue to the possible mechanistic basis of their signaling defects, as detailed in the Discussion. A majority (12 out of 17) of the receptors carrying suppressor changes alone also exhibited non-responsive-ON behavior (Figure 10). Two suppressor mutants exhibited locked-OFF outputs, and three responded to serine with moderate sensitivities, but very low cooperativities (Hill coefficients <5; wild-type Hill ~15).
Signaling Properties of Pseudorevertant Receptors
I utilized FRET-based kinase CheA assays to evaluate the signaling properties of the Tsr- F396G/W pseudorevertant receptors. In the host UU2567 (CheR-, CheB-), the F396G double mutants with suppressor changes in the flexible bundle region of Tsr were capable of modulating CheA activity in response to serine stimuli but had K1/2 values above that of wild-type Tsr, indicative of ON-shifted behavior (Figure 11).
In UU2567, F396G pseudorevertants with suppressor changes in the hairpin tip region were unresponsive to serine stimuli but exhibited kinase activity in the KCN assay (Figure 11). This behavior indicates a strong shift non-responsive shift toward a kinase-ON output state. It is important to note that these response data were collected in a host strain that lacked the adaptation enzymes (CheR-, CheB-). However, the doubly mutant receptors can support chemotaxis in an adaptation-competent (CheR+ CheB+) strain.
These results indicate that there may be more than one kinase-ON output state that enables these mutant receptors to be shifted toward the kinase-OFF output state. Continued work in the FRET reporter strain UU2700 will further elucidate the mechanism behind this model, as summarized in the Discussion.
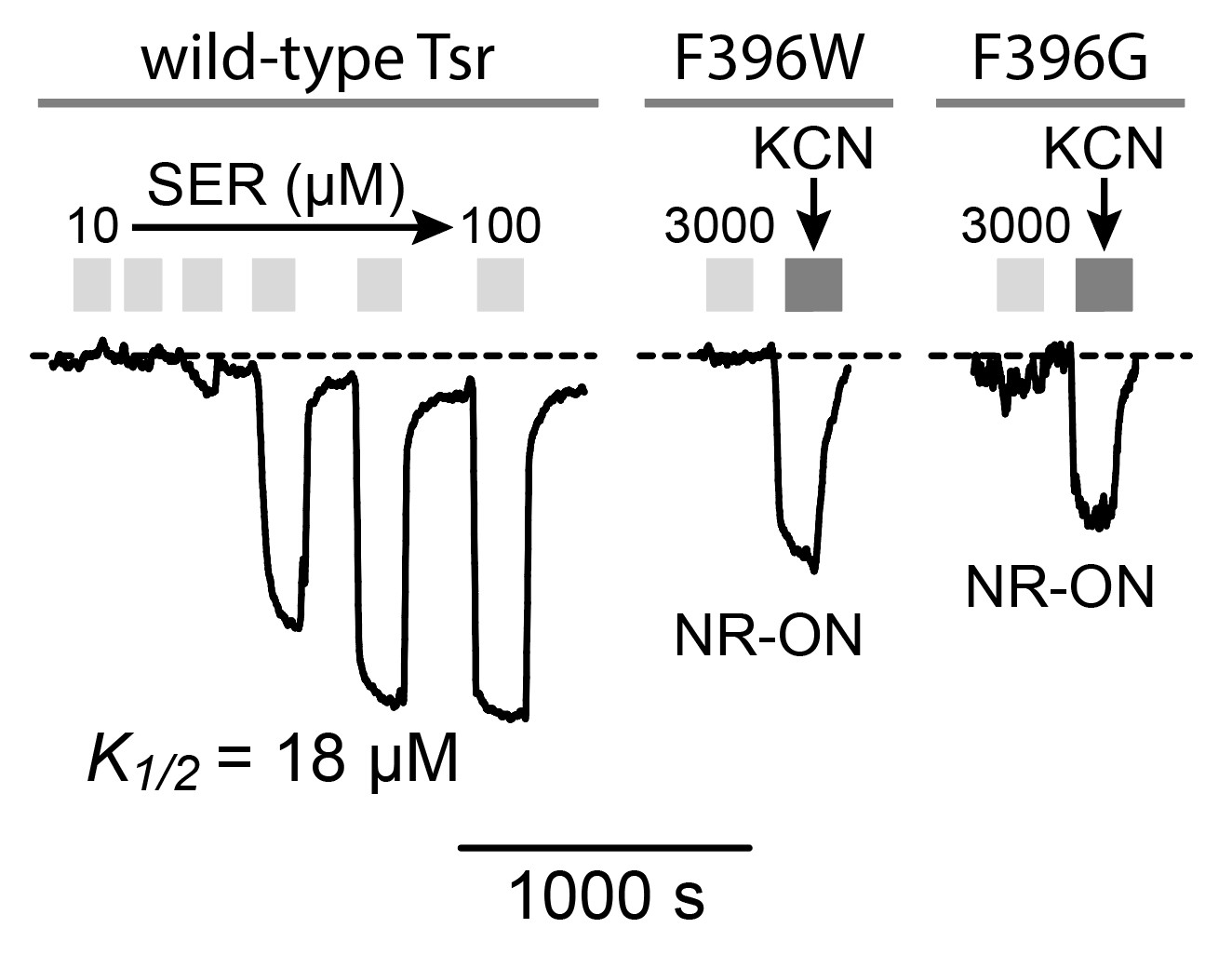
Figure 9: FRET kinase readouts. A. Example of wild-type Tsr FRET kinase readout. The y- axis represents the FRET signal which directly measures kinase activity (Sourjik et al, 2007). Light grey bars represent each time the receptor was introduced to serine. The wild-type receptor responds to serine and inhibits kinase activity. B. Dark grey bars represent kinase activity. Apparent kinase activity of mutant receptors F396G and F396W is not as high as the wild-type receptor.
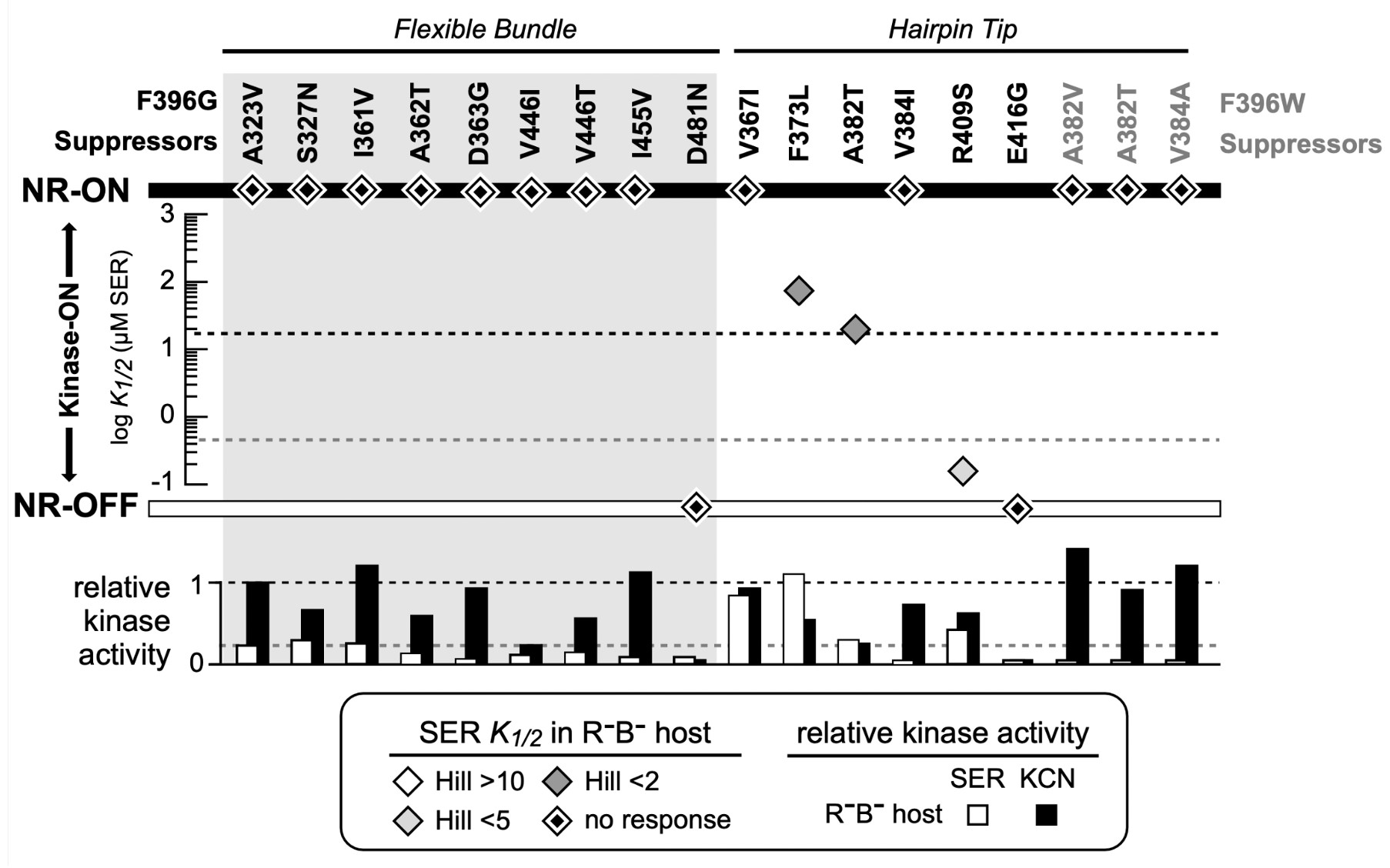
Figure 10: FRET Kinase assay of suppressor mutants for both Tsr-F396G and Tsr- F396W. The diamonds indicate the K1/2 measurement in the host UU2567 (CheR-, CheB-). Suppressor mutants that produce kinase activity but are unresponsive to serine are indicated by a diamond on the NR-ON bar. Suppressor mutants that are both unresponsive to serine and do not produce kinase activity are indicated by a diamond on the NR-OFF bar. The bottom graph depicts the relative kinase activity of the suppressor mutants in UU2567.
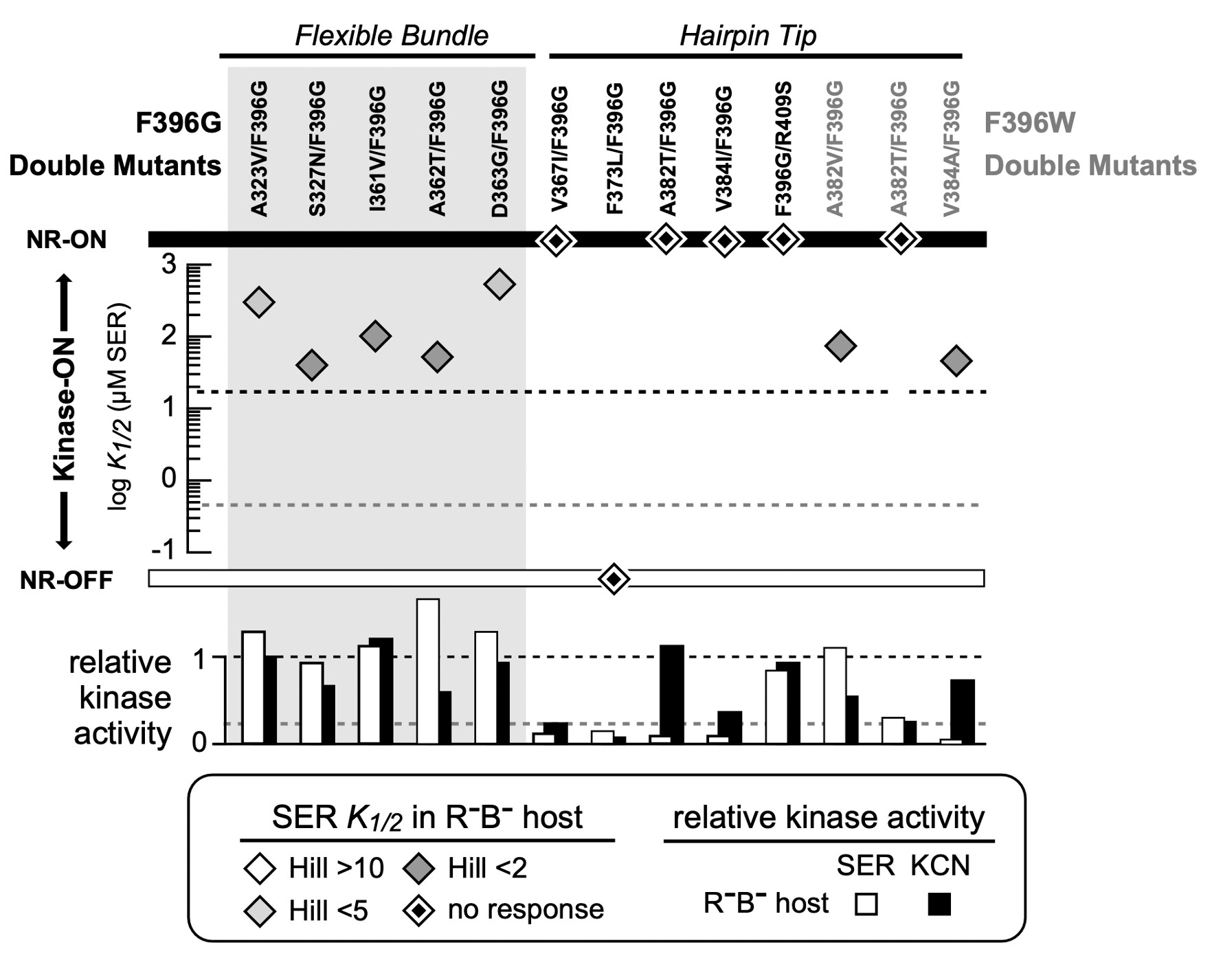
Figure 11: FRET Kinase assay of double mutants for both Tsr-F396G and Tsr-F396W mutants. The diamonds indicate the K1/2 measurement in the host UU2567 (CheR-, CheB-). Double mutants that produce kinase activity but are unresponsive to serine are indicated by a diamond on the NR-ON bar. Suppressor mutants that are both unresponsive to serine and do not produce kinase activity are indicated by a diamond on the NR-OFF bar. The bottom graph depicts the relative kinase activity of the suppressor mutants in UU2567.
DISCUSSION
In this project I isolated and characterized second-site mutations that effectively healed the Tsr-F396G and Tsr-F396W signaling defects. In the following, I will discuss these results in the context of a two-state receptor signaling model and suggest a suppression mechanism that involves dynamic signaling states at the cytoplasmic tip of the receptor. Moreover, the investigation of this position will uncover the mechanisms by which Tsr generates its locomotor- controlling signals and, in part, answer the question of how signaling is transmitted throughout the receptor–a pathway that is universal across organisms.
F396 as a switch for signal-state control
The current view of protein allostery hypothesizes that chemoreceptors such as Tsr occupy different conformations to modulate signal outputs. Inputs such as serine change the conformational landscape of the receptor to shift the hairpin tip to a kinase-OFF conformation (Ortega et al, 2013). In the absence of a chemical gradient, the Tsr receptor is mainly in a kinase- ON state.
F396 appears to play a central role in the ON-OFF conformational switch through its aromatic stacking interactions. One F396/F396’ stacking arrangement produces kinase-ON output; a flip in their stacking arrangement promotes an alternative interaction to shift Tsr to the kinase-OFF state (Ortega et al, 2013). This interaction occurs synchronously with signal conformations at the tip of the receptor. No other amino acid residue at Tsr residue 396 is able to promote wild-type chemotaxis. Many of such mutants are stuck in kinase-ON output, indicating that F396 is important for stabilizing the kinase-OFF confirmation of the receptor.
Suppression of F396 mutant signaling behaviors
To elucidate the mechanistic role that F396 plays, I hypothesized that second-site suppressors of F396G and F396W might act by enabling them to once again access the kinase- OFF state in response to serine stimuli. Six F396G suppressors and all three F396W suppressors altered hairpin tip residues (Figure 6). Of these, two F396G suppressors (F373L and V384I) were at trimer contact residues These results indicate that compensatory changes in the hairpin tip may enhance stability of the kinase-OFF output state. Nine F396G suppressors altered residues in the flexible region that adjoins the hairpin tip. These suppressor changes may alter the transmission of stimulus-induced conformational signals that modulate the structure of the hairpin tip.
The signaling properties of the parental F396G and F396W mutant receptors indicate that they might affect Tsr performance differently. I theorized that the F396W defect would be easier to heal because its aromatic side chain is very similar, albeit larger, than that of the wild-type phenylalanine. Interestingly, this was not the case. F396G pseudorevertants arose more readily and regained better chemotactic ability than did F396W double mutants. Perhaps the size of the amino acid sidechain at residue 396 is an important factor in its function. Although Tsr-F396W has an aromatic side chain similar to that of phenylalanine, its larger size might actively perturb the structure of the hairpin tip. Or, more likely, its large size makes switching between kinase-ON and kinase-OFF output states more difficult due steric hindrance.
Paradoxical signaling properties of Tsr mutant receptors
In the host UU2567, Tsr-F396G and Tsr-F396W failed to respond to any concentration of serine but were able to promote kinase activity (Figure 9). However, their kinase activity was only about half that of the native receptor. I expected the their suppressors might promote a kinase- OFF signaling behavior and thereby “offset” the locked-ON defects of the parental F396 mutant receptors.
Surprisingly, most of the suppressor changes created ON-shifted or locked-ON output behaviors. How might F396G (NR-ON) and its suppressors (NR-ON) create a double mutant capable of modulating serine? Perhaps there is a range of chemoreceptor conformations at the hairpin tip that elicit kinase activity over a number of dynamic states. Thus, there may exist more than one receptor ON state that can be driven toward a kinase-OFF output state by an attractant stimulus.
Operational conformations at the tip of the receptor
I present my mechanistic ideas in the context of a model where the receptor tip of Tsr can transition across a structural landscape characterized by different conformations and different dynamic behaviors. I propose that the receptor and its signaling hairpin tip operate across a variety of landscapes that define the kinase-ON and kinase-OFF output states (Figure 12A). As the receptor traverses these landscapes, its kinase activity will fluctuate. As such, the ON state of a mutant receptor might not be identical to the level of kinase activity in a wild-type receptor.
Suppose that the wild-type receptor (Tsr) operates within the shaded region of the conformational landscape shown in Figure 12B. In this regime, the attractant can interact with a receptor that is in the kinase-ON output state to drive it to kinase-OFF. Conversely, in the absence of the attractant, the native receptor will fluctuate between kinase-ON and kinase-OFF regions of the landscape. It is important to note that Tsr operates over a range of conformations that are conducive to control by the adaptation enzymes. The enzyme CheR operates on OFF-state receptors to shift them toward ON output by adding methyl groups to the receptor adaptation sites. Alternatively, the demethylating enzyme CheB operates on kinase-ON receptors and shifts them toward the OFF output state. In a two-state equilibrium the wild-type receptor can access both output states. However, if CheB is unable to operate on ON-state receptors that are too far outside of the normal wild-type range this would suggest that a two-state model of receptor signaling is too simplistic.
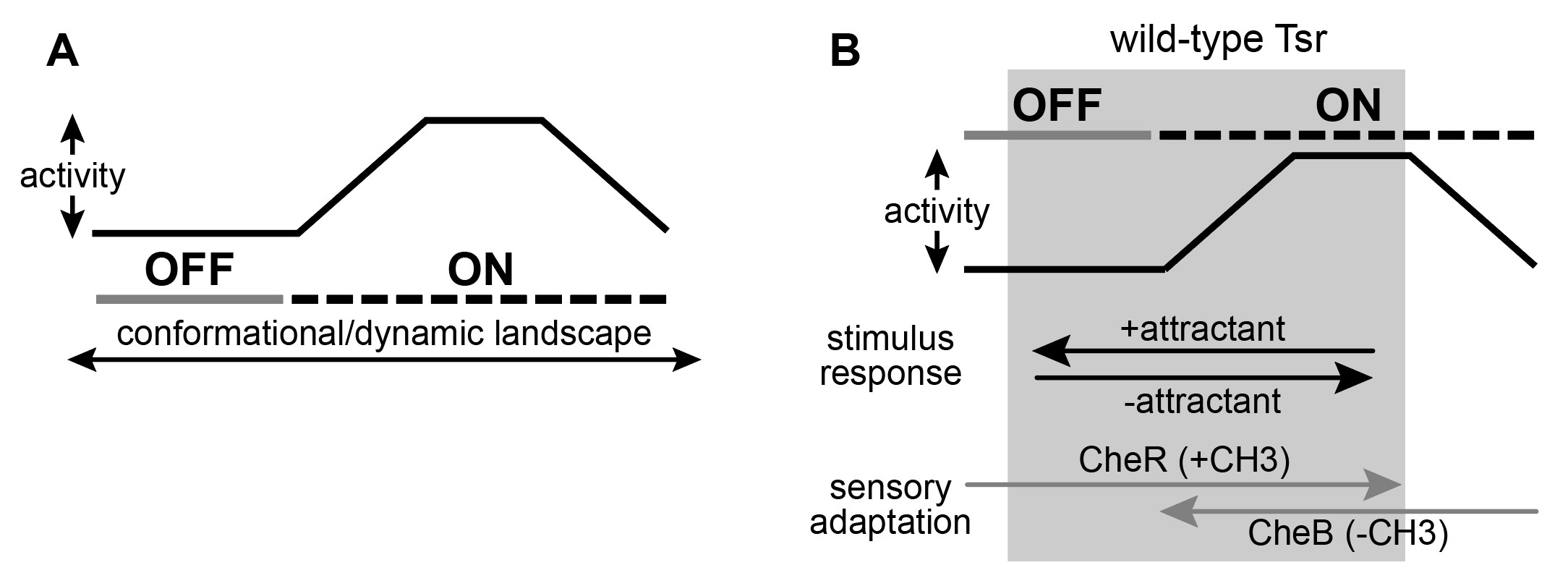
Figure 12: A graphical representation of the two-state model with dynamic “on” states. A. There may exist an operational landscape over which Tsr can transition by different conformations and differ dynamic behavior at the cytoplasmic tip which define the output states of kinase-on and kinase-off. B. Wild-type Tsr is works in the grey shaded region of the landscape and can shift between kinase-on and kinase-off states.
Figure 13 shows the conformational landscape of the Tsr-F396G receptor. As previously mentioned, F396G is constrained to a conformational landscape that produces less kinase activity than the native receptor and that cannot be driven OFF by serine (Ortega et al, 2013). To restore serine responsiveness, F396G must acquire a kinase-ON conformation similar to that of wild- type (Figure 13B).
I hypothesize that a suppressor of the F396G defect might itself be off-shifted and thus capable of shifting the doubly mutant receptor into a more wild-type conformational landscape. However, FRET kinase assays revealed that many of the found suppressor mutants did not respond to serine and produced less than the wild-type kinase activity. It seems as though the suppressor changes have constrained the kinase-ON conformational range of the receptor so that it is no longer capable of reaching the off-state. By contrast, in a low modification state, for example through CheB action, the Tsr-F396G can reach the serine-induced OFF state. Perhaps in combination, the F396 and suppressor defects interact in a way that shifts the conformational landscape of the doubly mutant receptor toward the wild-type operational range.
Evidence for different operational landscapes in different output states
Surprisingly, this interaction is both necessary and sufficient for proper transmembrane signing by Tsr pseudorevertant receptors. The conformational range of the doubly mutant receptor is much wider than that of the suppressor or of F396G separately. This could occur through two conformational interactions: one that “pulls” the F396G defect into a higher kinase activity range similar to that of wild-type Tsr, and one that confers serine-responsiveness to the suppressor defect. Thus, together the two defects create a receptor whose operational landscape is similar to the wild-type receptor and that supports chemotaxis.
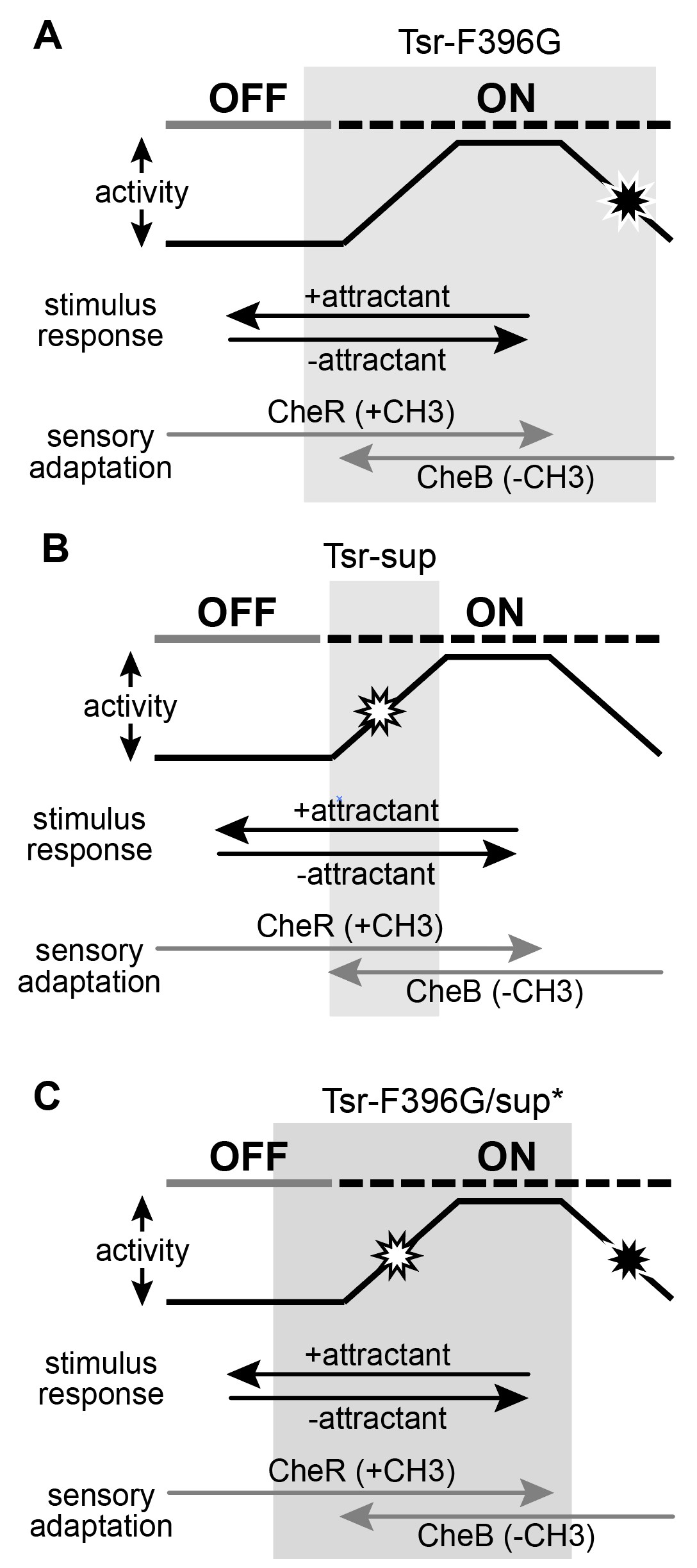
Figure 13: Conformational landscapes of mutant receptors. A. Tsr-F396G mutant receptor operates in the grey region of the landscape. It is not capable of reaching the kinase-off state without the adaptation enzyme CheB. B. Suppressor mutants have constricted the conformation range over which the receptor can operate, as shown by the grey bar. C. Double mutants have increased the conformational range over which the receptor can access both kinase-on and kinase-off states, shifting it into a landscape similar to the wild-type.
REFERENCES
Ames P, Parkinson JS. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine receptor. J Bacteriol 176:6340-6248.
Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9292-7. doi: 10.1073/pnas.0602135103. Epub 2006 Jun 2.
PMID: 16751275; PMCID: PMC1482603.
Ames P, Studdert CA, Reiser RH, Parkinson JS. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA 99:7060-7065. http://dx.doi.org/10.1073/pnas.092071899.
Ames, Peter, et al. “Collaborative Signaling by Mixed Chemoreceptor Teams in Escherichia Coli.” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no. 10, 2002, pp. 7060–65. JSTOR, http://www.jstor.org/stable/3058791. Accessed 30 Nov. 2022.
Ariane Briegel, Mark S Ladinsky, Catherine Oikonomou, Christopher W Jones, Michael J Harris, Daniel J Fowler, Yi-Wei Chang, Lynmarie K Thompson, Judith P Armitage, Grant J Jensen (2014) Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling eLife 3:e02151 https://doi.org/10.7554/eLife.02151
Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2015, Feb;72(4):691-708. doi: 10.1007/s00018-014-1770-5. Epub 2014 Nov 6. Review. PubMed PMID: 25374297.
Briand L, Marcion G, Kriznik A, Heydel JM, Artur Y, Garrido C, Seigneuric R, Neiers F. A self- inducible heterologous protein expression system in Escherichia coli. Sci Rep. 2016 Sep 9;6:33037. doi: 10.1038/srep33037. PMID: 27611846; PMCID: PMC5017159.
Chang ACY, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplamid. J Bacteriol 134:1141-1156
Kitanovic S, Ames P, Parkinson JS. A Trigger Residue for Transmembrane Signaling in the Escherichia coli Serine Chemoreceptor. J Bacteriol. 2015 Aug 1;197(15):2568-79. doi: 10.1128/JB.00274-15. Epub 2015 May 26. PMID: 26013490; PMCID: PMC4518824.
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4.
Nature. 1970 Aug 15;227(5259):680-5. doi: 10.1038/227680a0. PMID: 5432063.
Lai RZ, Han XS, Dahlquist FW, Parkinson JS. Paradoxical enhancement of chemoreceptor detection sensitivity by a sensory adaptation enzyme. Proc Natl Acad Sci U S A. 2017 Sep 5;114(36):E7583-E7591. doi: 10.1073/pnas.1709075114. Epub 2017 Aug 21. PMID: 28827352; PMCID: PMC5594695.
Lai RZ, Parkinson JS. 2014. Functional suppression of HAMP domain signaling defects in the E. coli
serine chemoreceptor. J Mol Biol 426:3642-3655 http://dx.doi.org/10.1016/j.jmb.2014.08.003.
Ortega, Davi R et al. “A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors.” Nature communications vol. 4 (2013): 2881. doi:10.1038/ncomms3881
Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015 May;23(5):257-66. doi: 10.1016/j.tim.2015.03.003. Epub 2015 Mar 30. PMID: 25834953; PMCID: PMC4417406.
Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol 151:106-113
Paulick A, Sourjik V. FRET Analysis of the Chemotaxis Pathway Response. Methods Mol Biol.
2018;1729:107-126. doi: 10.1007/978-1-4939-7577-8_11. PMID: 29429087.
Piñas G.E., DeSantis M.D., Cassidy C.K. and J.S. Parkinson (2022) Hexameric rings of the scaffolding protein CheW enhance response sensitivity and cooperativity in Escherichia coli chemoreceptor arrays. Sci. Signal., 15: DOI: 10.1126/scisignal.abj1737.
Piñas GE, DeSantis MD, Cassidy CK, Parkinson JS. Hexameric rings of the scaffolding protein CheW enhance response sensitivity and cooperativity in Escherichia coli chemoreceptor arrays. Sci Signal. 2022 Jan 25;15(718):eabj1737. doi: 10.1126/scisignal.abj1737. Epub 2022 Jan 25. PMID: 35077199; PMCID: PMC9261748.
Silversmith RE, Guanga GP, Betts L, Chu C, Zhao R, Bourret RB. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol. 2003 Mar;185(5):1495-502. doi: 10.1128/JB.185.5.1495-1502.2003. PMID: 12591865; PMCID: PMC148069.
Sourjik V, Berg HC. 2002. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A
99:123-127. http://dx.doi.org/10.1038/nature02406.
Sourjik V, Vaknin A, Shimizu TS, Berg HC. 2007. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol 423:365-391. http://dx.doi.org/10.1016/S0076- 6879(07)23017-4.
Stalla D, Akkaladevi N, White TA, Hazelbauer GL. Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity. Int J Mol Sci. 2019 Jun 19;20(12):2989. doi: 10.3390/ijms20122989. PMID: 31248079; PMCID: PMC6628036.
Yang W, Cassidy CK, Ames P, Diebolder CA, Schulten K, Luthey-Schulten Z, Parkinson JS, Briegel A. In Situ Conformational Changes of the Escherichia coli Serine Chemoreceptor in Different Signaling States. mBio. 2019 Jul 2;10(4):e00973-19. doi: 10.1128/mBio.00973-19. PMID: 31266867; PMCID: PMC6606802.
Zhou Q, Ames P, Parkinson JS. 2011. Biphasic control logic of HAMP domain signaling in the Escherichia coli serine chemoreceptor, Mol Microbiol 80:596-611. http://dx.doi.org/10.1111/j.1365-2958.2011.07577.x.

