Science
95 A Novel Organelle of the Synapse
Gareema Dhiman
Faculty Mentor: Erik Jorgensen (School of Biological Sciences, University of Utah)
During neurotransmission, neurons constantly fuse and recycle synaptic vesicles from the presynaptic membrane. Like other organelles, synaptic vesicles and proteins must be removed when damaged or overused. Build-up of damaged proteins may underlie neurodegenerative diseases like Parkinson’s Disease. However, a pathway for removal of synaptic material is unknown. Common degradative pathways of neuronal cells include the autophagy, proteasome, and endo-lysosomal systems. Damage within any of these pathways leads to accumulation of waste at synapses. Members of the Jorgensen lab have recently discovered a new organelle in Caenorhabditis elegans called a surveillant. The lysosome-derived organelle is found to probe synaptic spaces and take up synaptic proteins and aggregates. Surveillant numbers doubled when stressed using a 34° C heat shock for 4 hours. Surveillants share important similarities to lysosomes; however, surveillants are transported to and from synapses, unlike lysosomes, which reside in the cell soma. To better understand the origins of surveillants, I aim to answer the question, what is the molecular composition of surveillants? To address this, we have tagged synaptic vesicle proteins as well as autophagy and lysosomal proteins with fluorescent proteins to measure if they colocalize with surveillants. Identifying whether colocalization occurs is important in uncovering what pathway surveillants are involved in and how they can be differentiated from other degradative organelles. This research is important for broadening our understanding of how neurons respond to severe stress, with relevance to human diseases such as stroke, Alzheimer’s, and Parkinson’s Disease.
A special thanks is also extended to my father, whose encouragement, support, and generosity has given me the strength and perseverance to pursue and achieve my goals.
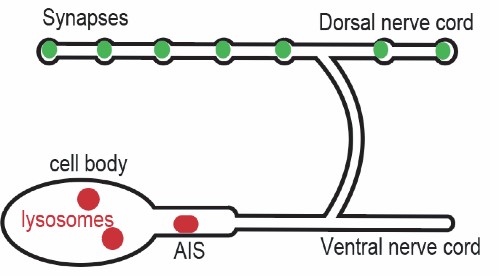
Figure 1: Lysosomes (red) are contained in the cell body and axon initial segment (AIS) in a C. elegans neuronal cell. At the synapses waste often builds up (green) that can lead to neurodegenerative diseases if not effectively degraded.
Proteins destined for degradation in the lysosome are transported by organelles such as late endosomes or autophagosomes. This allows for proper sorting and recycling of cellular components at synapses (Yu, 2018). During macroautophagy (generally referred to as autophagy) a double membrane forms around damaged proteins or other small organelles to form an autophagosome (Figure 2) (Yu, 2018). The autophagosomes are transported back along the axon and eventually fuse with lysosomes. In addition to macroautphagy, damaged proteins and organelles as well as endocytic vesicles can be degraded through microautophagy, endosomal microautophagy, and chaperone-mediated autophagy. Another main degradative pathway is the proteasome/ubiquitin (Figure 2). E3 ubiquitin ligases recognize and attach ubiquitin sequences to damaged proteins, which are cleaved into small peptides in the proteasome. Failure of any one of these pathways may lead to an accumulation of waste and cellular debris at synapses (Yu, 2018). Determining the precise mechanisms by which synaptic vesicles and proteins are recycled is therefore critical in the context of understanding neurodegenerative disease.
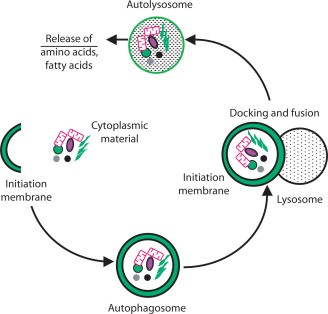
A B
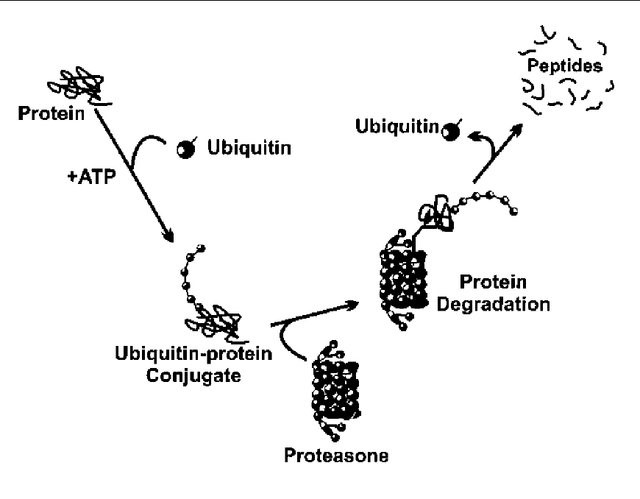
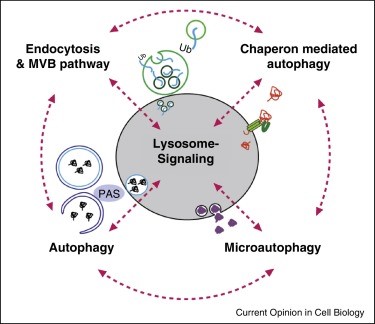
C
Figure 2:
(A) A closer depiction of the autophagy process from Wilfred, 2008. The process is initiated by the formation of a membrane that enlarges into a double membraned vesicle known as the autophagosome. This sequesters molecules and organelles. The autophagosome eventually fuses with a lysosome to form an autolysosome. The contents withing are degraded by acidic lysosomal hydrolase. (B) Overall pathways in which lysosomes are involved in from Lukas, 2016. Autophagy, endocytosis, multivescular bodies (MVB) pathway, chaperone mediated autophagy, and microautophagy are shown to be regulated by lysosomal signaling. (C) Basic proteasome pathway in protein degradation from Gheith, 2009. Proteins are first conjugated with ubiquitin by an ATP-dependent reaction. The protein is recognized and degraded to peptides by proteasome.
Environmental stresses such as heat shock, hypoxia, and starvation can directly affect lysosome biogenesis and function in C. elegans. Heat shock increases the number and size of lysosomes in neurons. In addition to an increase in lysosomes, the Jorgensen lab has observed the formation of a unique organelle called a surveillant (Wnukowski, 2023). The surveillant is likely derived from lysosomes, but unlike the lysosome can leave the AIS to “surveil” synapses, taking up protein aggregates. We present observations of possible proteins that may make up the molecular composition or are being taken up by surveillants in the synapse through a degradative pathway. Surveillants serve as a unique organelle that may be arise as a response to environmental stressors when the normal degradation pathways are overwhelmed.
BACKGROUNDC. ELEGANS AS A MODEL SYSTEM:
To determine the molecular components of surveillants, I performed studies using the nematode Caenorhabditis elegans (Figure 3).
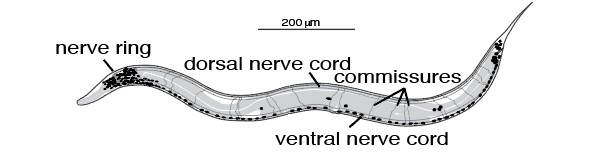
Figure 3: Caenorhabditis elegans diagram highlighting nerve cords and commissures.
C. elegans offer several advantages to work with to study surveillants: they require minimal labor in upkeep, have a rapid reproductive cycle (~3 days), and are self- fertilizing hermaphrodites. Males can be easily generated for genetic crossing of strains. C elegans are highly amenable to transgenesis and provides an ideal system to study co- localization of proteins due to its simple morphology, transparency, and annotated genome sequence (Sugi 2016). Additionally, many genes involved in autophagy, lysosomal degradation, and synaptic transmission are conserved from C. elegans to humans, which provides a simple model to understand the basic mechanisms of lysosomal function in the context of human diseases (Sugi, 2016).
LYSOSOME LABELING
LMP-1 (an ortholog of human LAMP2 [lysosomal associated membrane protein 2]) is a known marker of lysosomes and an abundant protein within the lysosome membrane (Settembre, 2013). LMP-1 can also be found on endosomes and transport organelles (Cheng, 2018). Generally, it can be difficult to label lysosomes due to the ambiguity in the definition of a lysosome. Some studies call all Lamp-1 labeled organelles lysosomes, while others describe lysosomes based on luminal pH (Liao, 2019; Bucci, 2000). Due to the low expression of lysosomes in neurons, knowledge regarding degradation through this pathway at synapses is scarce. Additionally, it can be difficult to assess whether lysosomes experience anterograde motion to axonal and synaptic spaces. In GABA neurons, the C. elegans kinesin adaptor UNC-16 restricts the transport of lysosomes past the AIS, blocking them from reaching the axon and synapse (Ferguson, 2017).
The Jorgensen lab has defined an organelle called a surveillant, which has lysosome like properties, but undergoes anterograde transport to axonal and synaptic spaces in neurons. To study surveillants by microscopy, single copy insertions of lysosomal and synaptic proteins fused to fluorescent proteins were expressed in C. elegans GABA neurons, driven by the unc-47 promoter. GABA neuron cell bodies are located in the ventral cord but send axons to the dorsal cord. By imaging only fluorescent proteins in the dorsal cord we can distinguish these organelles from somatic lysosomes.
LGG-1
LGG-1 within C. elegans enables GABA receptor binding and ubiquitin protein ligase binding. It is also known as an autophagosome marker which has been used throughout this experiment. Using LGG-1 has allowed for colocalization experiments with surveillants to be done and further understand how the organelle differs from autophagosomes.
SURVEILLANTS
Using a tandem fluorescent reporter, surveillants are described as mCherry::LGG-1 labeled organelles in the dorsal cord and are differentiated from autophagosomes by a quenching of eGFP::LGG-1 (Figure 4). Under acidic conditions, eGFP is degraded in lysosomes, whereas mCherry is acid resistant, suggesting surveillants are acidified like lysosomes. mCherry::LGG-1 organelles can bud off somatic lysosomes, suggesting these are surveillants of a lysosomal origin.
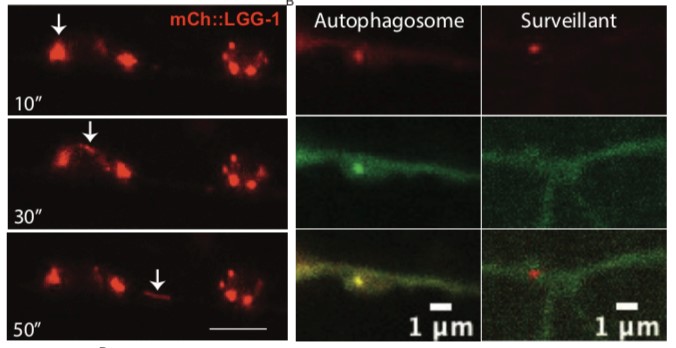
Figure 4: (A) mCherry::LGG-1 surveillants are seen budding off from a cell body. Numbers on bottom left represent time in seconds. (B) Image of an autophagosome in the dorsal nerve cord and a surveillant. Autophagosomes are seen labeled with GFP while surveillants are labeled by mCherry. Figure courtesy of Christine Wnukowski.
Typically, synaptic proteins at axons and presynaptic terminals undergo retrograde transport for degradation by somatic lysosomes (Cheng, 2015; Okerlund, 2017), which typically remain constrained to the AIS. Uniquely, surveillants are seen leaving the AIS and travelling to the dorsal cord where the synapses are located (Figure 5, Figure 6). The organelle is seen to have bidirectional movement, undergoing both anterograde and retrograde movement. Surveillants pause at synapses to “surveil” the area and take up synaptic cargo (Wnukowski, 2023). The organelle has been detected taking up α- synuclein which accumulates at neurons in Parkinson’s.
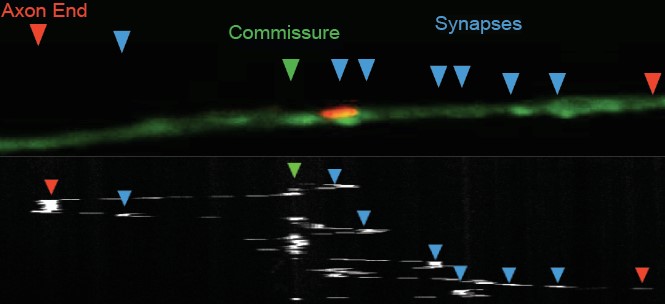
Figure 5: The kymograph confirms the movements of surveillants that have been seen. After the surveillant exited the AIS it visits the dorsal cord and moves back and forth, spending some time there. Figure courtesy of Christine Wnukowski.
Synapses
Dorsal nerve cord
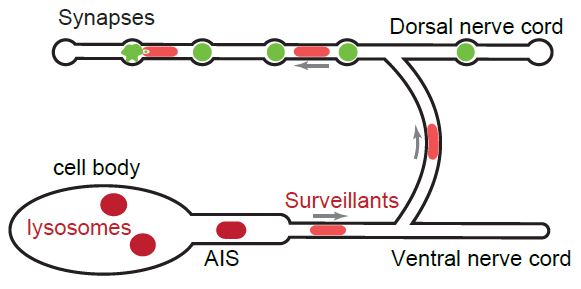
Figure 6: Depiction of the unique movement seen by surveillants (light red) going past the AIS, up to the dorsal cord and synapses. As the surveillants move to the synapse they may be picking up waste or cargo (green) to degrade.
The molecular makeup and cargo that is picked up by surveillants remains unknown. Therefore, I investigated the co-localization of unique organelle markers with surveillants to understand their identity and origin.
METHODSTABLE 1: STRAINS
| Name | Strain | Proteins Colocalized |
| Wild-type | N2 | wild-type |
| EG10058 | LGG-1(oxSi1131[Punc-47::mCh::gfp::lgg-1::let-858]) IV | LGG-1 Tandem Fluorescent |
| EG10057 | oxSi784[Punc-47::GFP::snap-29::let-858utr, CBunc-119+] II oxSi1110[Punc-47::mCh::lgg-1::let-858, CB-unc-119(+)] | SNAP-29/LGG-1 |
| UNC-32 | 5605(oxSi1097[PSnt-1::2xnls-FLP(G5D) unc-119+] ) II
ox904[unc-32::flpon-gfp] III
oxSi1110[Punc-47::mCh::lgg-1::let-858, CB-unc-119(+)] IV |
UNC-32/LGG-1 |
| RAB-7 | 5605(oxSi1097[PSnt-1::2xnls-FLP(G5D) unc-119+] ) II
rab-7(ox984)[Skylan-S::FLP::rab-7] II
oxSi1110[Punc-47:mCherry::lgg-1::let-858, CB-unc-119(+)] IV |
RAB-7/LGG-1 |
| CUP-15 | ox980[cup-15::FLP-on::eGFP] II
oxEx2241[Psnt1:2xNLS-FLP(G5D), Psnt-1:mTagBFP2, Pmyo2:eGFP, unc-119(+)]
oxSi1110[Punc-47:mCherry::lgg-1::let-858, CB-unc-119(+)] IV |
CUP-15/LGG-1 |
Table 1: List of C. elegan strains used throughout the experiment. RAB-7 and CUP-15 have preliminary data on them which is further addressed in the Discussion section.
WORM CARE
Each strain was maintained by transferring worms to new plates twice a week. The plates contained food made up of the E. coli strain, OP50. This food source is known as an uracil auxotroph with limited growth on Nematode Growth Medium (NGM). The plates used OP50 and spread over NGM plates.
When transferring the worms to a new plate, a worm picker was used. This consists of a 1 inch 32-gauge platinum wire mounted onto a Pasteur pipet. The wire is used to pick the worms under a microscope by swiping the side of the worm and can be sanitized between transfers by being flamed. Lastly, the worms were consistently stored at 15°C.
MOUNTING & HEATSHOCK
To stage the worms as young adults, L4 worms were picked onto a fresh plate the day before imaging. Worms were heat shocked for 3 hours in a 34°C incubator, or kept at 15°C as a control. Worms were then allowed to rest at room temperature for 15 minutes after heat shock, followed by mounting and imaging.
Worms were placed on a 2% agarose, pad mounted on a glass slide into 2 µl of 180 mM muscimol and 2 µl of 30 µm polystyrene beads. A #1.5 coverslip was placed on top and sealed with petroleum jelly to prevent dehydration. The agarose allowed for the worms to stick onto the slide and the muscimol was used to inhibit muscle contractions through interactions with the GABAA receptor UNC-49 (Han, 2015). As a result of this the worm relaxes and becomes immobile. The polystyrene beads provide additional movement restriction by allowing contact with the glass (worms are ~ 50 µm in diameter) while preventing the worms from being fully compressed.
IMAGING PROCESS
CONFOCAL AND AIRYSCAN MICROSCOPY
Confocal and Airyscan imaging was performed on a Zeiss 880 Line Scanning Microscope. Zeiss software was used to capture optical sections in the Z direction. Images were obtained using a Plan-Apochromat 63x/1.4 NA Oil immersion objective. Standard laser lines and filter sets were used to capture mCherry and GFP signals.
Skylan-S images were collected using GFP lasers and filters along with 0.5% illumination using a 405 nm laser to switch it to the “on” state. Surveillants were identified by scanning the dorsal nerve cord and commissures for Punc-47::mCherry::lgg- 1 positive structures.
IMAGEJ AND COLOCALIZATION SCORING
Colocalization was manually calculated in FIJI/ImageJ. Areas with a surveillant were deemed a region of interest (ROI) based on fluorescence intensity. The ROI was then used to measure fluorescence intensity of the second fluorescent protein. If the ROI was twice as bright as the background intensity, it was considered colocalized.
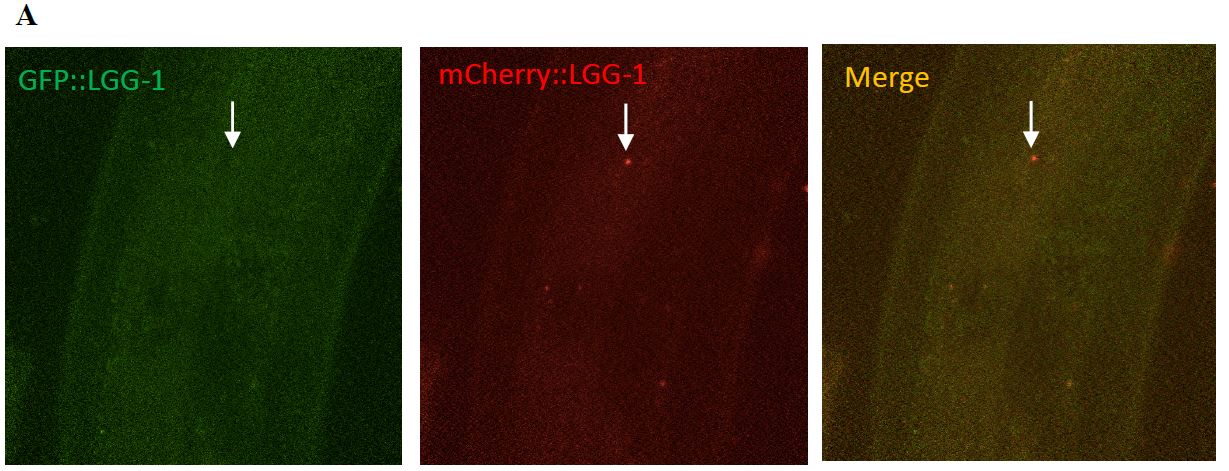
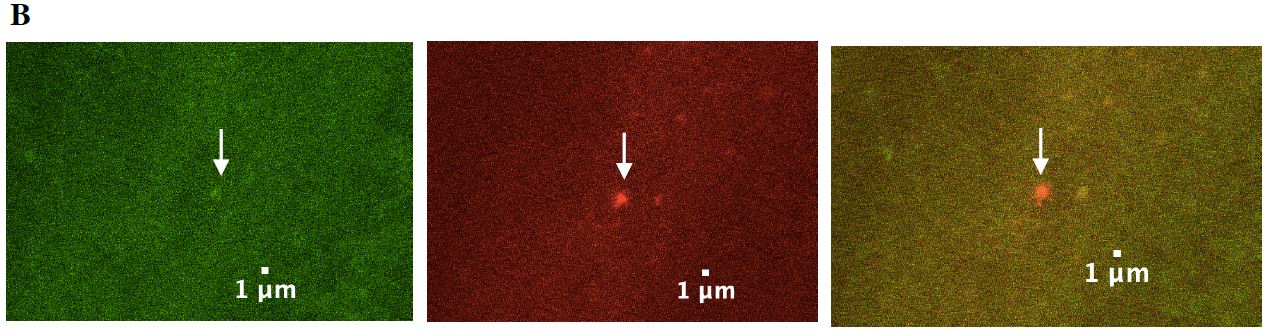
Figure 7: A surveillant seen moving through the commissures. (A) The top three images depict surveillants not labeled in the green channel but in the red channel. This indicates that surveillants are not labeled by the autolysosome marker, GFP and is labeled by mCherry. (B) The lower three images are a zoomed in look at the images above.
COLOCALIZATION DATA UNC-32
UNC-32 is an ortholog of the a subunit of the V0 sector of the V-ATPase and is highly
expressed in C. elegans neurons. UNC-32 is required for acidifying organelles and is present on both lysosomes and synaptic vesicles. UNC-32 was found on 2/11 (18%) of surveillants, suggesting that surveillants do not require UNC-32 (Table 2), (Figure 8, Figure 9). This observation was important in distinguishing the makeup of surveillants from proteins that are commonly found to be associated with lysosomes.
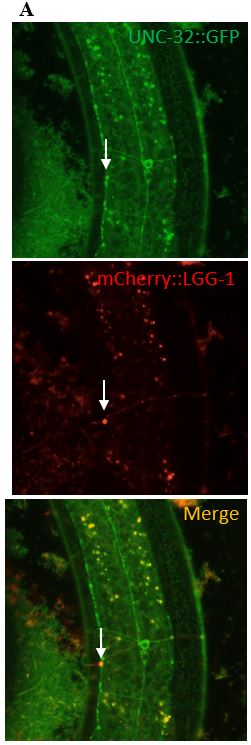

Figure 8: (A) The images in the left column depict what a clear colocalization of UNC-32 and a surveillant looks like. The surveillant was seen travelling down the commissure of the worm.
(B) The images in the right column are images of a surveillant that did not colocalize with UNC-
32. The surveillant was seen travelling down a commissure but is not associated with anything seen in the green channel.
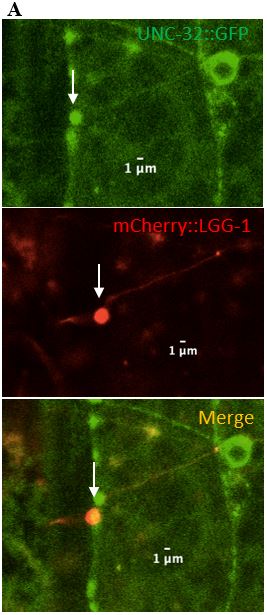
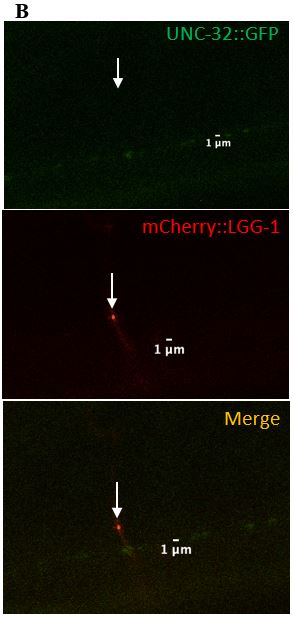
Figure 9: (A) Zoomed in version of the colocalized surveillant shown in Figure 8, column A (B) Zoomed in version of the surveillant that did not colocalize with UNC-32 from Figure 8, column B.
SNAP-29
SNAP-29 is a member of a SNARE complex, and helps fuse autophagosomes to lysosomes, forming an autolysosome. SNAP-29 colocalizes with mCherry puncta in 14/22 (64%) surveillants. This indicates that surveillants are similar to or may come from autolysosomes (Figure 10, Figure 11).
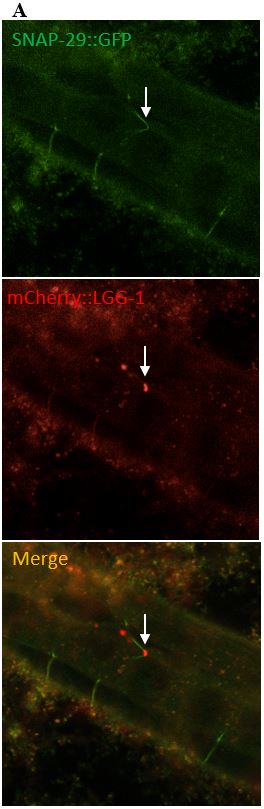
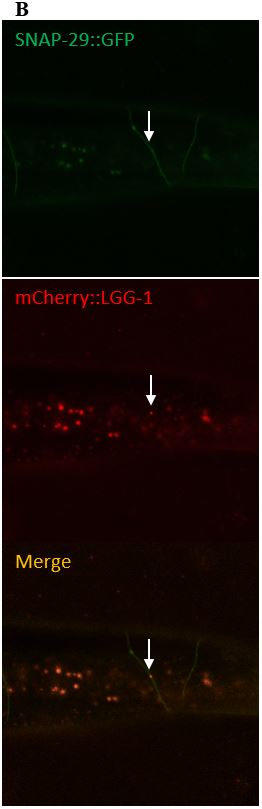
Figure 10: (A) The images on the left depict a surveillant that colocalized with SNAP-29 within a commissure. (B) The images in the column on the right depicts a surveillant also seen within a commissure but did not colocalize with SNAP-29.
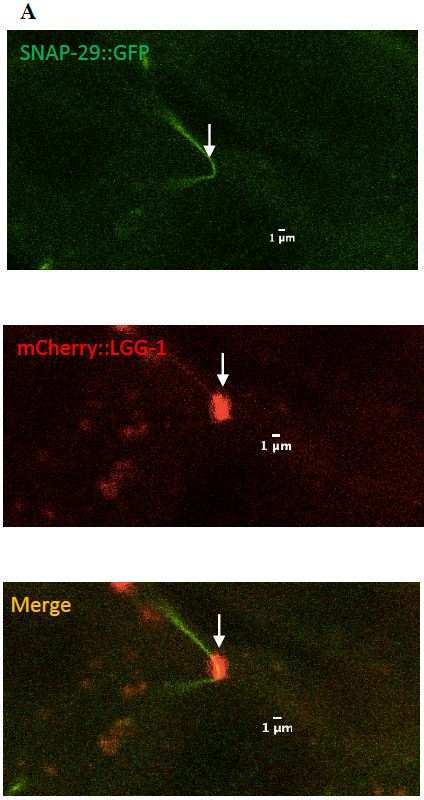
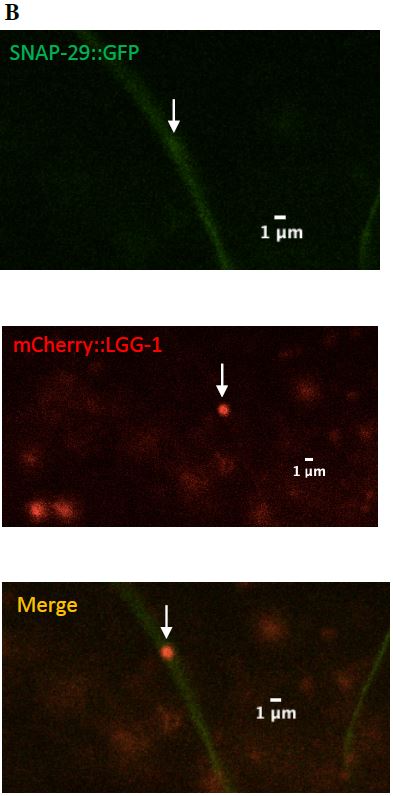
Figure 11: (A) Zoomed in version of the colocalized surveillant shown in Figure 9, column A
(B) Zoomed in version of the surveillant that did not colocalize with SNAP-29 from Figure 9, column B.
TABLE 2: COLOCALIZATION RESULTS
| Protein | Amount Colocalized | Amount not Colocalized | Total number of Surveillants
Identified |
Percent Colocalized |
| GFP::LGG-
1/mCherry::LGG-1 |
0 | 17 | 17 | 0% |
| UNC-32/LGG-1 | 2 | 9 | 11 | 18% |
| SNAP-29/LGG-1 | 14 | 8 | 22 | 64% |
DISCUSSIONDisruptions in degradative pathways and accumulation of aggregated proteins are often hallmarks of common neurodegenerative diseases such as Parkinson’s disease. How these damaged and aggregated proteins are removed from synapses is still unclear. We have observed that surveillants are likely derived from autolysosomes and may be a unique pathway by which neurons respond to stress.
Surveillants have been detected at synapses using the mCherry::LGG-1 marker. This also indicates that surveillants are distinct from autophagosomes and autolysosomes due to the quenching of eGFP::LGG-1, suggesting an acidic environment.
LIMITATIONS
The heat shock process used in this experiment is essential in detecting surveillants due to their scarcity under basal conditions. Although the heat shock acting as a stressor to the worms may be a large contributor to the visibility of surveillants, mCherry could potentially contribute as lysosomal stressor (Costantini, 2015; Landgraf, 2013). mCherry may be accumulating in lysosomes because it is resistant to lysosomal proteases (Melentijevic, 2017; Shemiakina, 2012). Additionally, eGFP variants are quenched in acidic environments, making it difficult to identity what proteins are colocalizing with surveillants. Difficulties in labeling lysosomes was another limitation within this experiment. Due to the ambiguous definition of a lysosome there are several distinct markers that can be used. We chose LC3 / LGG-1 since it is also known as an autophagosome marker and allows us to better differentiate and understand the composition of surveillants from other degradative organelles.
FUTURE DIRECTIONS
We currently see that surveillants will take up alpha-synuclein, a protein that is mutated and aggregates in Parkinson’s disease. I have also seen that surveillants have colocalized with the proteins RAB-7 and CUP-15. In the future, I would like to look at surveillant co- localization with RAB-7 (a late endosome/lysosome marker), and the lysosome chloride transporter osteoporosis-associated transmembrane protein 1 OSTM1/CUP-15 (a true lysosomal marker). First, RAB-7 is of interest because while the Jorgensen lab has identified that surveillants are not lysosomes, endosomes have the ability to be transported into the axon beyond the AIS, like surveillants. Second, CUP-15 is a specific marker of lysosomes because it has no known association with other organelles. Both genes have been tagged with fluorescent proteins using CRISPR/Cas9. Using the FRT/Flp system, we can induce expression of these fluorescent proteins specifically in neurons. Another possible protein found in surveillants is the trans Golgi network protein TGN-38. SNAP-29 is associated with TGN-38 that acts as a trans-Golgi shuttle protein. Currently, TGN-38 has been seen to be present on all surveillants. However, when this experiment was done, the protein was overexpressed using an array. To validate colocalization of TGN-38 and surveillants a CRISPR that endogenously tags the protein will have to be generated. This preliminary data suggests surveillants are lysosome-derived organelles that could be transporting or degrading synaptic cargo. The research on surveillants has been important in broadening our understanding of how waste could be removed from neurons to prevent neurodegenerative diseases and how human neuron’s respond to severe stress.
REFERENCES1. Alzheimer’s Association. 2023 Alzheimer’s Disease Facts and Figures. Alzheimers Dement 2023;19(4). DOI 10.1002/alz.13016.
2. Bucci, C., Thomsen, P., Nicoziani, P., Mccarthy, J., & Deurs, B. Van. (2000). Rab7: a key to lysosome biogenesis. Molecular Biology of the Cell, 11(February), 467–480.
3. Cheng, X. T., Zhou, B., Lin, M. Y., Cai, Q., & Sheng, Z. H. (2015). Axonal Autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. Journal of Cell Biology, 209(3), 377–386.
4. Cheng, Xiu-Tang & Xie, Yuxiang & Zhou, Bing & Huang, Ning & Farfel- Becker, Tamar & Sheng, Zu-Hang. (2018). Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. The Journal of Cell Biology. 217. jcb.201711083. 10.1083/jcb.201711083.
5. Costantini, L. M., Baloban, M., Markwardt, M. L., Rizzo, M., Guo, F., Verkhusha, V. V., & Snapp, E. L. (2015). A palette of fluorescent proteins optimized for diverse cellular environments. Nature Communications, 6(May). https://doi.org/10.1038/ncomms8670
6. Di Maio, Roberto & Barrett, Paul & Hoffman, Eric & Barrett, Caitlyn & Zharikov, Alevtina & Borah, Anupom & Hu, Xiaoping & McCoy, Jennifer & Chu, Charleen & Burton, Edward & Hastings, Teresa & Greenamyre, J.. (2016). – Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinsons disease. Science Translational Medicine. 8. 342ra78-342ra78. 10.1126/scitranslmed.aaf3634.
7. Edwards, S. L., Yu, S. C., Hoover, C. M., Phillips, B. C., Richmond, J. E., & Miller, K. G. (2013). An organelle gatekeeper function for caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics, 194(1), 143–161. https://doi.org/10.1534/genetics.112.147348
8. Ferguson, S. M. (2017a). Neuronal lysosomes. Neuroscience Letters, 697(October 2017), 1–9. https://doi.org/10.1016/j.neulet.2018.04.005
9. Gheith, Osama & Wafa, Ehab & Refaie, Ayman & Hassan, Nabil & Ismail, Amani & Sheashaa, Hussein & Shokeir, Ahmaed & Kamal, Mohamed & Ghoneim, Mohamed. (2009). Post-transplant anemia in pediatric patients and its impact on patient and graft survival: single center experience. African Journal of Nephrology. 13. 31-38. 10.21804/13-1-777.
10. Han B, Bellemer A, Koelle MR. An evolutionarily conserved switch in response to GABA affects development and behavior of the locomotor circuit of Caenorhabditis elegans. Genetics. 2015 Apr;199(4):1159-72. doi: 10.1534/genetics.114.173963. Epub 2015 Feb 2. PMID: 25644702; PMCID:
PMC4391577.https://doi.org/10.1016/j.neuron.2017.01.026
11. Kundra R, Dobson CM, Vendruscolo M. A Cell- and Tissue-Specific Weakness of the Protein Homeostasis System Underlies Brain Vulnerability to Protein Aggregation. iScience. 2020 Mar 27;23(3):100934. doi:10.1016/j.isci.2020.100934. Epub 2020 Feb 24. PMID: 32146327; PMCID: PMC7063235.
12. Landgraf, D., Akumus, B., Chien, P., Baker, T. A., & Paulsson, J. (2013). Segregation of molecules at cell division reveals native protein localization. Nature Methods, 9(5), 480–482. https://doi.org/10.1038/nmeth.1955.Segregation
13. Liao, Y. C., Fernandopulle, M. S., Wang, G., Choi, H., Hao, L., Drerup, C. M., Patel, R., Qamar, S., Nixon-Abell, J., Shen, Y., Meadows, W., Vendruscolo, M., Knowles, T. P. J., Nelson, M., Czekalska, M. A., Musteikyte, G., Gachechiladze,M. A., Stephens, C. A., Pasolli, H. A., … Ward, M. E. (2019). RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell, 179(1), 147-164.e20. https://doi.org/10.1016/j.cell.2019.08.050
14. Lie, P. P. Y., Yang, D. S., Stavrides, P., Goulbourne, C. N., Zheng, P., Mohan, P. S., Cataldo, A. M., & Nixon, R. A. (2021). Post-Golgi carriers, not lysosomes, confer lysosomal properties to pre-degradative organelles in normal and dystrophic axons. Cell Reports, 35(4), 109034. https://doi.org/10.1016/j.celrep.2021.109034
15. Lukas A Huber, David Teis, Lysosomal signaling in control of degradation pathways, Current Opinion in Cell Biology, Volume 39, 2016, Pages 8-14, ISSN 0955-0674, https://doi.org/10.1016/j.ceb.2016.01.006.
16. Melentijevic, I., Toth, M. L., Arnold, M. L., Guasp, R. J., Harinath, G., Nguyen, K. C., Taub, D., Parker, J. A., Neri, C., Gabel, C. V., Hall, D. H., & Driscoll, M. (2017). C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature, 542(7641), 367–371. https://doi.org/10.1038/nature21362
17. Monaco, Antonio & Fraldi, Alessandro. (2020). Protein Aggregation and Dysfunction of Autophagy-Lysosomal Pathway: A Vicious Cycle in Lysosomal Storage Diseases. Frontiers in Molecular Neuroscience. 13. 10.3389/fnmol.2020.00037.
18. Okerlund, N. D., Schneider, K., Leal-ortiz, S., Montenegro-venegas, C., Kim, S. A., Garner, L. C., Gundelfinger, E. D., Reimer, R. J., & Garner, C. C. (2017).Bassoon controls presynaptic autophagy through Atg5. Neuron, 93(4), 897- 913.e7.
19. Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009 Feb;19(2):52-61. doi: 10.1016/j.tcb.2008.11.002. Epub 2008 Dec 26. PMID: 19112021; PMCID: PMC4843516.
20. Recasens, Ariadna & Dehay, Benjamin & Bové, Jordi & Carballo-Carbajal, Iria & Dovero, Sandra & Pérez, A & Fernagut, Pierre-Olivier & Blesa, Javier & Parent, Annabelle & Perier, Celine & Farinas, Isabel & Obeso, Jose & Bezard, Erwan & Vila, Miquel. (2014). Lewy body extracts from Parkinson disease brains trigger synuclein pathology and neurodegeneration in mice and monkeys. Annals of neurology. 75. 10.1002/ana.24066.
21. Settembre, C., Fraldi, A., Medina, D. L., & Ballabio, A. (2013). Signals from the lysosome : a control centre for cellular clearance and energy metabolism. Nature Publishing Group, 14(5), 283–296. https://doi.org/10.1038/nrm3565
22. Shemiakina, I. I., Ermakova, G. V., Cranfill, P. J., Baird, M. A., Evans, R. A., Souslova, E. A., Staroverov, D. B., Gorokhovatsky, A. Y., Putintseva, E. V., Gorodnicheva, T. V., Chepurnykh, T. V., Strukova, L., Lukyanov, S., Zaraisky, A. G., Davidson, M. W., Chudakov, D. M., & Shcherbo, D. (2012). A monomeric red fluorescent protein with low cytotoxicity. Nature Communications, 3, 1–7. https://doi.org/10.1038/ncomms2208
23. Sugi T. Genome Editing in C. elegans and Other Nematode Species. Int J Mol Sci. 2016 Feb 26;17(3):295. doi: 10.3390/ijms17030295. PMID: 26927083; PMCID: PMC4813159.
24. Wilfred Lieberthal, (2008), Macroautophagy: a mechanism for mediating cell death or for promoting cell survival?, Kidney International, Volume 74, Issue 5, Pages 555-557, ISSN 0085-2538, https://doi.org/10.1038/ki.2008.325.
25. Wnukowski, C., (2023). Neuronal lysosomes are dynamically managed in number, size, localization, and mobility in response to stress. [Unpublished chapter of doctoral dissertation]. University of Utah.
26. Yu, L., Chen, Y., & Tooze, S. A. (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy, 14(2), 207–215. https://doi.org/10.1080/15548627.2017.1378838
Internet Resources:
- WormBase
- – Available from: https://wormbase.org/
- FP Base
- – Available from: https://fpbase.org/
- Wormbook
- – Available from: http://www.wormbook.org/

