85 Protein Structure
LumenLearning
Protein Structure
Each successive level of protein folding ultimately contributes to its shape and therefore its function.
LEARNING OBJECTIVES
Summarize the four levels of protein structure
KEY TAKEAWAYS
Key Points
- Protein structure depends on its amino acid sequence and local, low-energy chemical bonds between atoms in both the polypeptide backbone and in amino acid side chains.
- Protein structure plays a key role in its function; if a protein loses its shape at any structural level, it may no longer be functional.
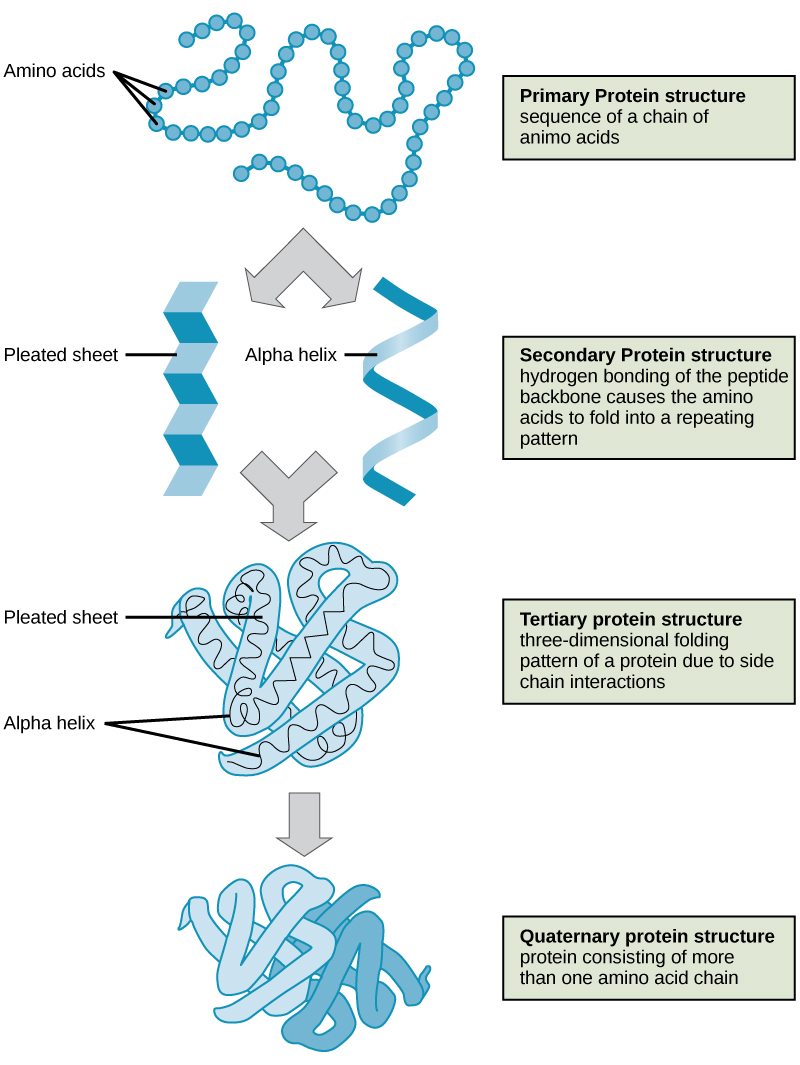
- Primary structure is the amino acid sequence.
- Secondary structure is local interactions between stretches of a polypeptide chain and includes α-helix and β-pleated sheet structures.
- Tertiary structure is the overall the three-dimension folding driven largely by interactions between R groups.
- Quarternary structures is the orientation and arrangement of subunits in a multi-subunit protein.
Key Terms
- antiparallel: The nature of the opposite orientations of the two strands of DNA or two beta strands that comprise a protein’s secondary structure
- disulfide bond: A bond, consisting of a covalent bond between two sulfur atoms, formed by the reaction of two thiol groups, especially between the thiol groups of two proteins
- β-pleated sheet: secondary structure of proteins where [latex]\text{N-H}[/latex] groups in the backbone of one fully-extended strand establish hydrogen bonds with [latex]\text{C=O}[/latex] groups in the backbone of an adjacent fully-extended strand
- α-helix: secondary structure of proteins where every backbone [latex]\text{N-H}[/latex] creates a hydrogen bond with the [latex]\text{C=O}[/latex] group of the amino acid four residues earlier in the same helix.
The shape of a protein is critical to its function because it determines whether the protein can interact with other molecules. Protein structures are very complex, and researchers have only very recently been able to easily and quickly determine the structure of complete proteins down to the atomic level. (The techniques used date back to the 1950s, but until recently they were very slow and laborious to use, so complete protein structures were very slow to be solved.) Early structural biochemists conceptually divided protein structures into four “levels” to make it easier to get a handle on the complexity of the overall structures. To determine how the protein gets its final shape or conformation, we need to understand these four levels of protein structure: primary, secondary, tertiary, and quaternary.
Primary Structure
A protein’s primary structure is the unique sequence of amino acids in each polypeptide chain that makes up the protein. Really, this is just a list of which amino acids appear in which order in a polypeptide chain, not really a structure. But, because the final protein structure ultimately depends on this sequence, this was called the primary structure of the polypeptide chain. For example, the pancreatic hormone insulin has two polypeptide chains, A and B.
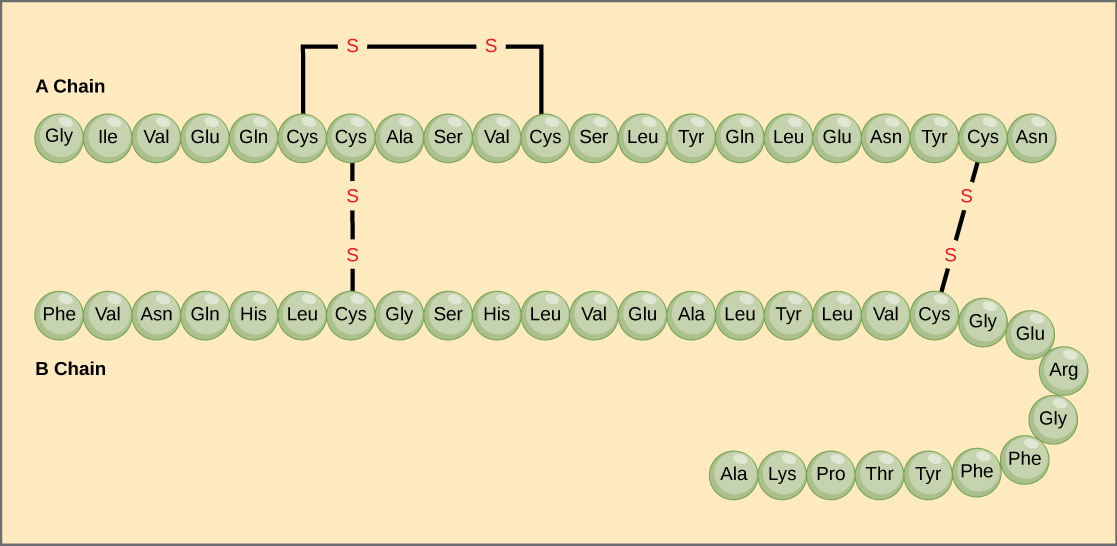
The gene, or sequence of DNA, ultimately determines the unique sequence of amino acids in each peptide chain. A change in nucleotide sequence of the gene’s coding region may lead to a different amino acid being added to the growing polypeptide chain, causing a change in protein structure and therefore function.
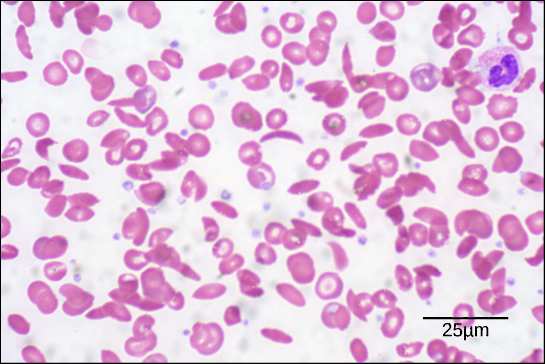
The oxygen-transport protein hemoglobin consists of four polypeptide chains, two identical α chains and two identical β chains. In sickle cell anemia, a single amino substitution in the hemoglobin β chain causes a change the structure of the entire protein. When the amino acid glutamic acid is replaced by valine in the β chain, the polypeptide folds into an slightly-different shape that creates a dysfunctional hemoglobin protein. So, just one amino acid substitution can cause dramatic changes. These dysfunctional hemoglobin proteins, under low-oxygen conditions, start associating with one another, forming long fibers made from millions of aggregated hemoglobins that distort the red blood cells into crescent or “sickle” shapes, which clog arteries. People affected by the disease often experience breathlessness, dizziness, headaches, and abdominal pain.
Secondary Structure
A protein’s secondary structure is whatever regular structures arise from interactions between neighboring or near-by amino acids as the polypeptide starts to fold into its functional three-dimensional form. Secondary structures arise as H bonds form between local groups of amino acids in a region of the polypeptide chain. Rarely does a single secondary structure extend throughout the polypeptide chain. It is usually just in a section of the chain. The most common forms of secondary structure are the α-helix and β-pleated sheet structures and they play an important structural role in most globular and fibrous proteins.
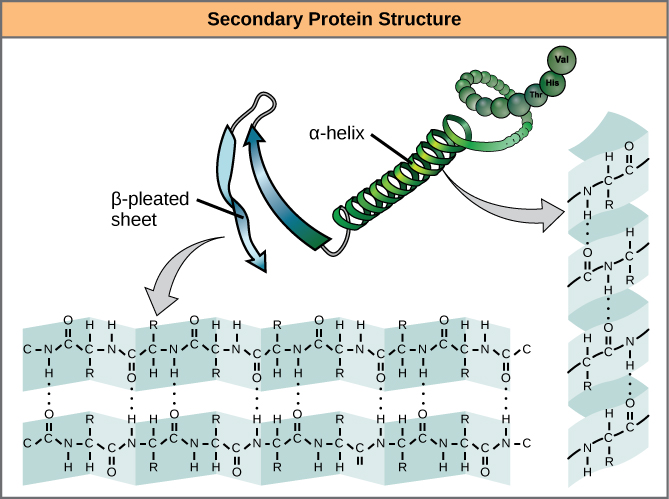
In the α-helix chain, the hydrogen bond forms between the oxygen atom in the polypeptide backbone carbonyl group in one amino acid and the hydrogen atom in the polypeptide backbone amino group of another amino acid that is four amino acids farther along the chain. This holds the stretch of amino acids in a right-handed coil. Every helical turn in an alpha helix has 3.6 amino acid residues. The R groups (the side chains) of the polypeptide protrude out from the α-helix chain and are not involved in the H bonds that maintain the α-helix structure.
In β-pleated sheets, stretches of amino acids are held in an almost fully-extended conformation that “pleats” or zig-zags due to the non-linear nature of single [latex]\text{C-C}[/latex] and [latex]\text{C-N}[/latex] covalent bonds. β-pleated sheets never occur alone. They have to held in place by other β-pleated sheets. The stretches of amino acids in β-pleated sheets are held in their pleated sheet structure because hydrogen bonds form between the oxygen atom in a polypeptide backbone carbonyl group of one β-pleated sheet and the hydrogen atom in a polypeptide backbone amino group of another β-pleated sheet. The β-pleated sheets which hold each other together align parallel or antiparallel to each other. The R groups of the amino acids in a β-pleated sheet point out perpendicular to the hydrogen bonds holding the β-pleated sheets together, and are not involved in maintaining the β-pleated sheet structure.
Tertiary Structure
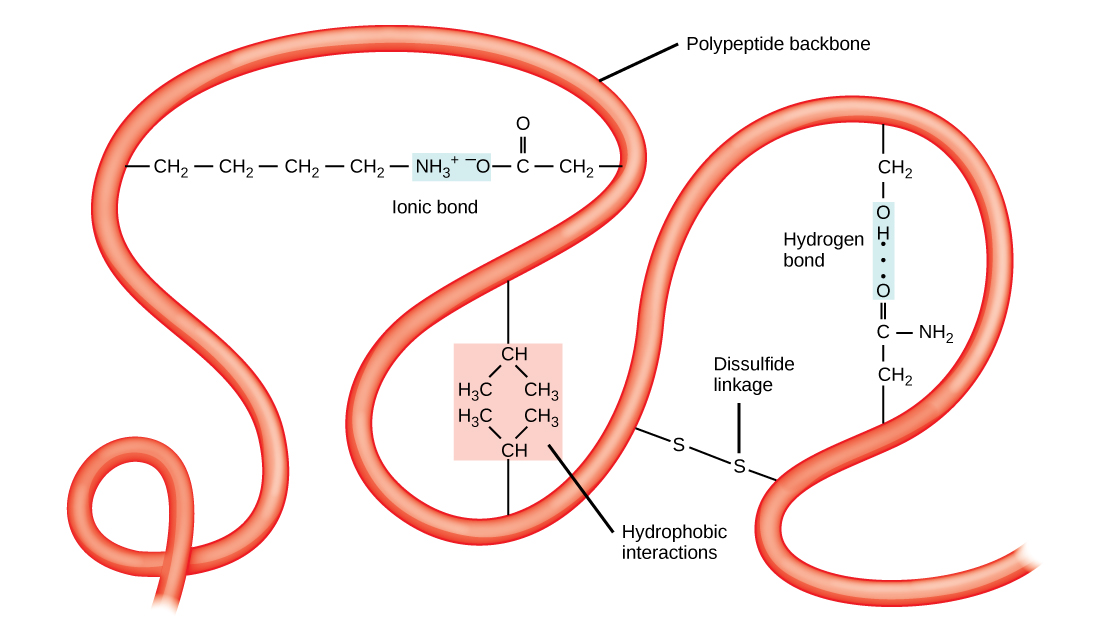
The tertiary structure of a polypeptide chain is its overall three-dimensional shape, once all the secondary structure elements have folded together among each other. Interactions between polar, nonpolar, acidic, and basic R group within the polypeptide chain create the complex three-dimensional tertiary structure of a protein. When protein folding takes place in the aqueous environment of the body, the hydrophobic R groups of nonpolar amino acids mostly lie in the interior of the protein, while the hydrophilic R groups lie mostly on the outside. Cysteine side chains form disulfide linkages in the presence of oxygen, the only covalent bond forming during protein folding. All of these interactions, weak and strong, determine the final three-dimensional shape of the protein. When a protein loses its three-dimensional shape, it will no longer be functional.
Quaternary Structure
The quaternary structure of a protein is how its subunits are oriented and arranged with respect to one another. As a result, quaternary structure only applies to multi-subunit proteins; that is, proteins made from more than one polypeptide chain. Proteins made from a single polypeptide will not have a quaternary structure.
In proteins with more than one subunit, weak interactions between the subunits help to stabilize the overall structure. Enzymes often play key roles in bonding subunits to form the final, functioning protein.
For example, insulin is a ball-shaped, globular protein that contains both hydrogen bonds and disulfide bonds that hold its two polypeptide chains together. Silk is a fibrous protein that results from hydrogen bonding between different β-pleated chains.
Denaturation and Protein Folding
Denaturation is a process in which proteins lose their shape and, therefore, their function because of changes in pH or temperature.
LEARNING OBJECTIVES
Discuss the process of protein denaturation
KEY TAKEAWAYS
Key Points
- Proteins change their shape when exposed to different pH or temperatures.
- The body strictly regulates pH and temperature to prevent proteins such as enzymes from denaturing.
- Some proteins can refold after denaturation while others cannot.
- Chaperone proteins help some proteins fold into the correct shape.
Key Terms
- chaperonin: proteins that provide favorable conditions for the correct folding of other proteins, thus preventing aggregation
- denaturation: the change of folding structure of a protein (and thus of physical properties) caused by heating, changes in pH, or exposure to certain chemicals
Each protein has its own unique sequence of amino acids and the interactions between these amino acids create a specify shape. This shape determines the protein’s function, from digesting protein in the stomach to carrying oxygen in the blood.
Changing the Shape of a Protein
If the protein is subject to changes in temperature, pH, or exposure to chemicals, the internal interactions between the protein’s amino acids can be altered, which in turn may alter the shape of the protein. Although the amino acid sequence (also known as the protein’s primary structure) does not change, the protein’s shape may change so much that it becomes dysfunctional, in which case the protein is considered denatured. Pepsin, the enzyme that breaks down protein in the stomach, only operates at a very low pH. At higher pHs pepsin’s conformation, the way its polypeptide chain is folded up in three dimensions, begins to change. The stomach maintains a very low pH to ensure that pepsin continues to digest protein and does not denature.
Enzymes
Because almost all biochemical reactions require enzymes, and because almost all enzymes only work optimally within relatively narrow temperature and pH ranges, many homeostatic mechanisms regulate appropriate temperatures and pH so that the enzymes can maintain the shape of their active site.
Reversing Denaturation
It is often possible to reverse denaturation because the primary structure of the polypeptide, the covalent bonds holding the amino acids in their correct sequence, is intact. Once the denaturing agent is removed, the original interactions between amino acids return the protein to its original conformation and it can resume its function.
However, denaturation can be irreversible in extreme situations, like frying an egg. The heat from a pan denatures the albumin protein in the liquid egg white and it becomes insoluble. The protein in meat also denatures and becomes firm when cooked.

Chaperone proteins (or chaperonins ) are helper proteins that provide favorable conditions for protein folding to take place. The chaperonins clump around the forming protein and prevent other polypeptide chains from aggregating. Once the target protein folds, the chaperonins disassociate.
LICENSES AND ATTRIBUTIONS
CC LICENSED CONTENT, SHARED PREVIOUSLY
- Curation and Revision. Provided by: Boundless.com. License: CC BY-SA: Attribution-ShareAlike
CC LICENSED CONTENT, SPECIFIC ATTRIBUTION
- hydrogen bond. Provided by: Wiktionary. Located at: http://en.wiktionary.org/wiki/hydrogen_bond. License: CC BY-SA: Attribution-ShareAlike
- OpenStax College, Biology. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/?collection=col11448/latest. License: CC BY: Attribution
- Boundless. Provided by: Boundless Learning. Located at: http://www.boundless.com//biology/definition/antiparallel. License: CC BY-SA: Attribution-ShareAlike
- disulfide bond. Provided by: Wiktionary. Located at: http://en.wiktionary.org/wiki/disulfide_bond. License: CC BY-SA: Attribution-ShareAlike
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_09.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_07.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_04.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_06.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_08.jpg. License: CC BY: Attribution
- OpenStax College, Biology. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/?collection=col11448/latest. License: CC BY: Attribution
- Structural Biochemistry/Cell Organelles/Cytoskeleton. Provided by: Wikibooks. Located at: http://en.wikibooks.org/wiki/Structural_Biochemistry/Cell_Organelles/Cytoskeleton%23Eukaryotic_cytosolic_chaperonin. License: CC BY-SA: Attribution-ShareAlike
- denaturation. Provided by: Wiktionary. Located at: http://en.wiktionary.org/wiki/denaturation. License: CC BY-SA: Attribution-ShareAlike
- chaperonin. Provided by: Wikipedia. Located at: http://en.wikipedia.org/wiki/chaperonin. License: CC BY-SA: Attribution-ShareAlike
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_09.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_07.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_04.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_06.jpg. License: CC BY: Attribution
- OpenStax College, Proteins. October 16, 2013. Provided by: OpenStax CNX. Located at: http://cnx.org/content/m44402/latest/Figure_03_04_08.jpg. License: CC BY: Attribution
- Protein Denaturation. Provided by: Wikipedia. Located at: http://en.wikipedia.org/wiki/File:Protein_Denaturation.png. License: CC BY-SA: Attribution-ShareAlike
This chapter is an adaptation of the chapter "Protein Structure" in Boundless Chemistry by LumenLearning and is licensed under a CC BY-SA 4.0 license.

