Transcription
Elizabeth Rebarchik
Objective 2: Describe transcription: how DNA is made into RNA.
Molecular Biology Objective 2 Video Lecture
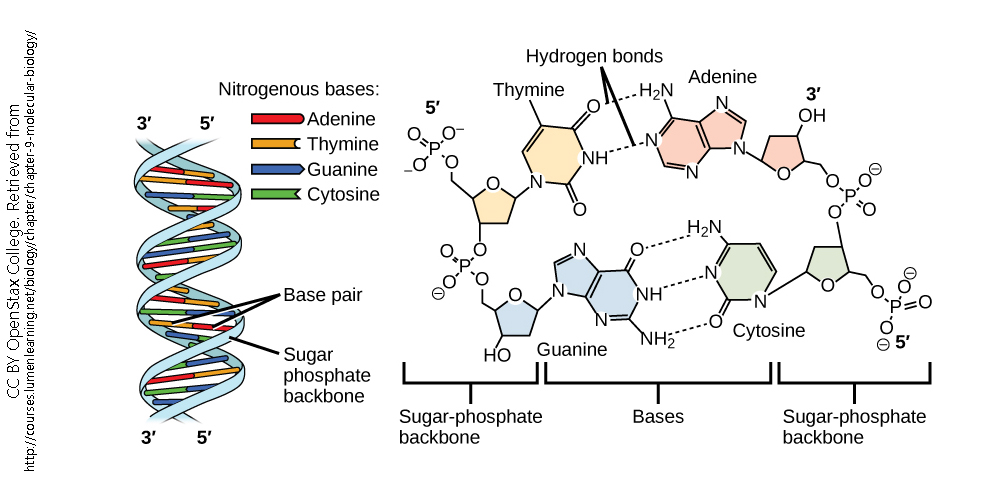 The structure of nucleic acids makes transcription possible. Recall that each nucleotide consists of:
The structure of nucleic acids makes transcription possible. Recall that each nucleotide consists of:
- a base (adenine, or A; cytosine, or C; guanine, or G; thymine, or T, found only in DNA; and uracil, or U, found only in RNA)
- a sugar (deoxyribose in DNA or ribose in RNA)
- a phosphate (PO43–) group
These are arranged in order from the 5′ (“five-prime”) end of the molecule to the 3′ (“three-prime”) end of the molecule. All operations on nucleic acid, including transcription, occur in the 5′ to 3′ direction.
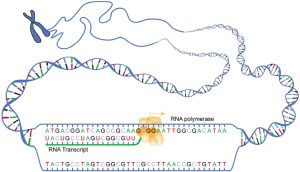
The enzyme used to make RNA from a DNA template is called RNA polymerase. This is a great name for this enzyme, because it makes a polymer out of RNA nucleotides (ribonucleotides).
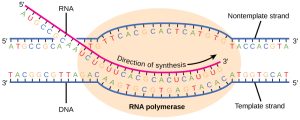 Transcription takes place in the nucleus, where DNA is packaged and stored. Reference books never leave the library. DNA never leaves the nucleus. The double-stranded DNA is transcribed to single-stranded RNA, with the A–T, C–G, G–C, and T–A base pairs of DNA coding for A, C, G and U bases along an RNA backbone. Remember that thymine in DNA is replaced by the very similar base uracil in RNA.
Transcription takes place in the nucleus, where DNA is packaged and stored. Reference books never leave the library. DNA never leaves the nucleus. The double-stranded DNA is transcribed to single-stranded RNA, with the A–T, C–G, G–C, and T–A base pairs of DNA coding for A, C, G and U bases along an RNA backbone. Remember that thymine in DNA is replaced by the very similar base uracil in RNA.
All cells have the same DNA, but not all cells have the same protein. How is this accomplished? It must be that some genes are switched “on” while others are switched “off”. For example, both your intestinal cells and your brain cells have the same DNA, but for intestinal cells we need to switch on enzymes and other proteins that are used to break down and move nutrients across the cell, while in brain cells we need to switch on enzymes and other proteins that are used to send signals from one cell to another.
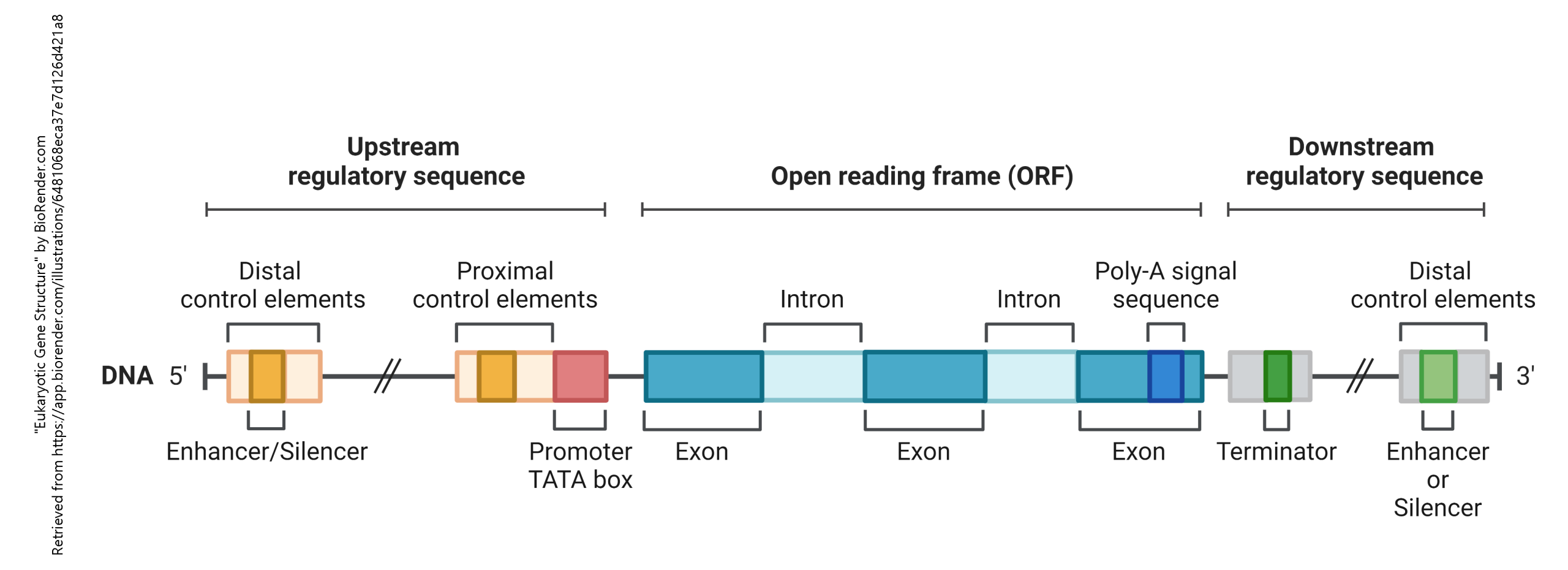 The switching “on” or “off” of genes occurs because of transcription factors that bind to regions of the DNA sequence ahead of (or sometimes behind) the RNA being made from the DNA template. The gene itself (i.e. the DNA which will be translated into protein) is called the open reading frame (ORF). Ahead of the ORF is the upstream regulatory sequence. In this part of the gene are enhancers, silencers, and promoters. All of these elements are found upstream (i.e. on the 5′ side) of the ORF. There is also a downstream regulatory sequence. This sequence of bases is found at the 3′ end of the gene. These are not nearly as well understood as the upstream regulatory sequences.
The switching “on” or “off” of genes occurs because of transcription factors that bind to regions of the DNA sequence ahead of (or sometimes behind) the RNA being made from the DNA template. The gene itself (i.e. the DNA which will be translated into protein) is called the open reading frame (ORF). Ahead of the ORF is the upstream regulatory sequence. In this part of the gene are enhancers, silencers, and promoters. All of these elements are found upstream (i.e. on the 5′ side) of the ORF. There is also a downstream regulatory sequence. This sequence of bases is found at the 3′ end of the gene. These are not nearly as well understood as the upstream regulatory sequences.
The RNA that codes for a protein is called messenger RNA (mRNA) because it carries a message to the apparatus that makes protein. mRNA is distinguished by the presence of 100-250 riboadenosine residues — a poly(A) tail — at the 3′ end of the message, part of the distal control elements shown in green and gray in the above diagram. If it doesn’t code for a protein, it’s called non-coding RNA (ncRNA) and there are two of these we’ll talk about (ribosomal RNA and transfer RNA) and a whole bunch we won’t talk about that have enzymatic activity even though they’re not proteins (ribozymes).
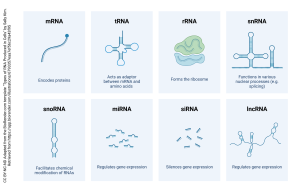
The RNA made by RNA polymerase and destined to code for protein is called pre-mRNA or the primary transcript. The primary transcript has to be edited to its final form before it is shipped out of the nucleus. The final edited form, which is shipped out of the nucleus, is what is formally called mRNA.
The exon is the portion of the DNA that is expressed, or made into protein.
The intron is the portion of the DNA that is not made into protein and must be edited out.
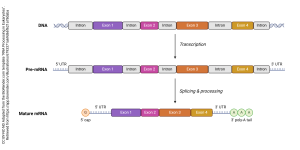
The process of transcribing from a segment of DNA (a gene) is shown here). The introns or intervening regions are edited out (grey regions). Exons are spliced together to make the final mRNA. This mRNA will be read on a ribosome and translated to a protein. We will study the process of translation in a following objective.
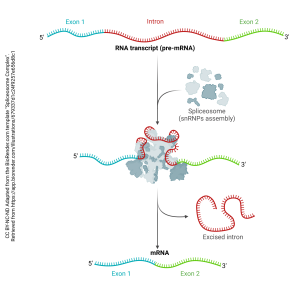
In order to create mRNA, the introns must be sliced out and the exons stitched together. This is accomplished by an organelle called the spliceosome, which is made up of several small nuclear ribonucleoprotein particles, or snRNPs (“snurps”). A structure called a lariat is formed, the intron is cut out, and the ends of the exon are stitched together.
This snRNP participates in the editing process that cuts and splices pre-mRNA into mRNA.
The edited transcript (now officially called mRNA) passes through a pore in the nuclear envelope. Once in the cytoplasm, it is used as the blueprint for protein synthesis at the ribosome protein factory.
An “average” chromosome has 150 million base pairs. Every so often, a string of base pairs about 100,000 nucleotides long will form a gene. There are about 1,000 genes on a chromosome, so that means only about 0.7% of the chromosome is used for genes.
Media Attributions
- DNA structure © Betts, J. Gordon; Young, Kelly A.; Wise, James A.; Johnson, Eddie; Poe, Brandon; Kruse, Dean H. Korol, Oksana; Johnson, Jody E.; Womble, Mark & DeSaix, Peter adapted by Jim Hutchins is licensed under a CC BY (Attribution) license
- Transcription © National Human Genome Research Institute is licensed under a Public Domain license
- Transcription © Samantha Fowler, Rebecca Roush, & James Wise is licensed under a CC BY (Attribution) license
- Eukaryotic gene structure © Catherine C is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Types of RNA Produced in Cells © Sally Kim adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- RNA Processing in Eukaryotes © BioRender adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Spliceosome Complex © BioRender adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license

