An Introduction to Some Molecular Biological Techniques Used in Neuroscience
Jim Hutchins
Objective 6: Cite some examples of the use of molecular biological techniques in neuroscience.
Molecular Biology Objective 6 Video Lecture
In many cases, neuroscientists would like to measure the amount of mRNA transcript which is being made in a particular cell or tissue at a particular time.
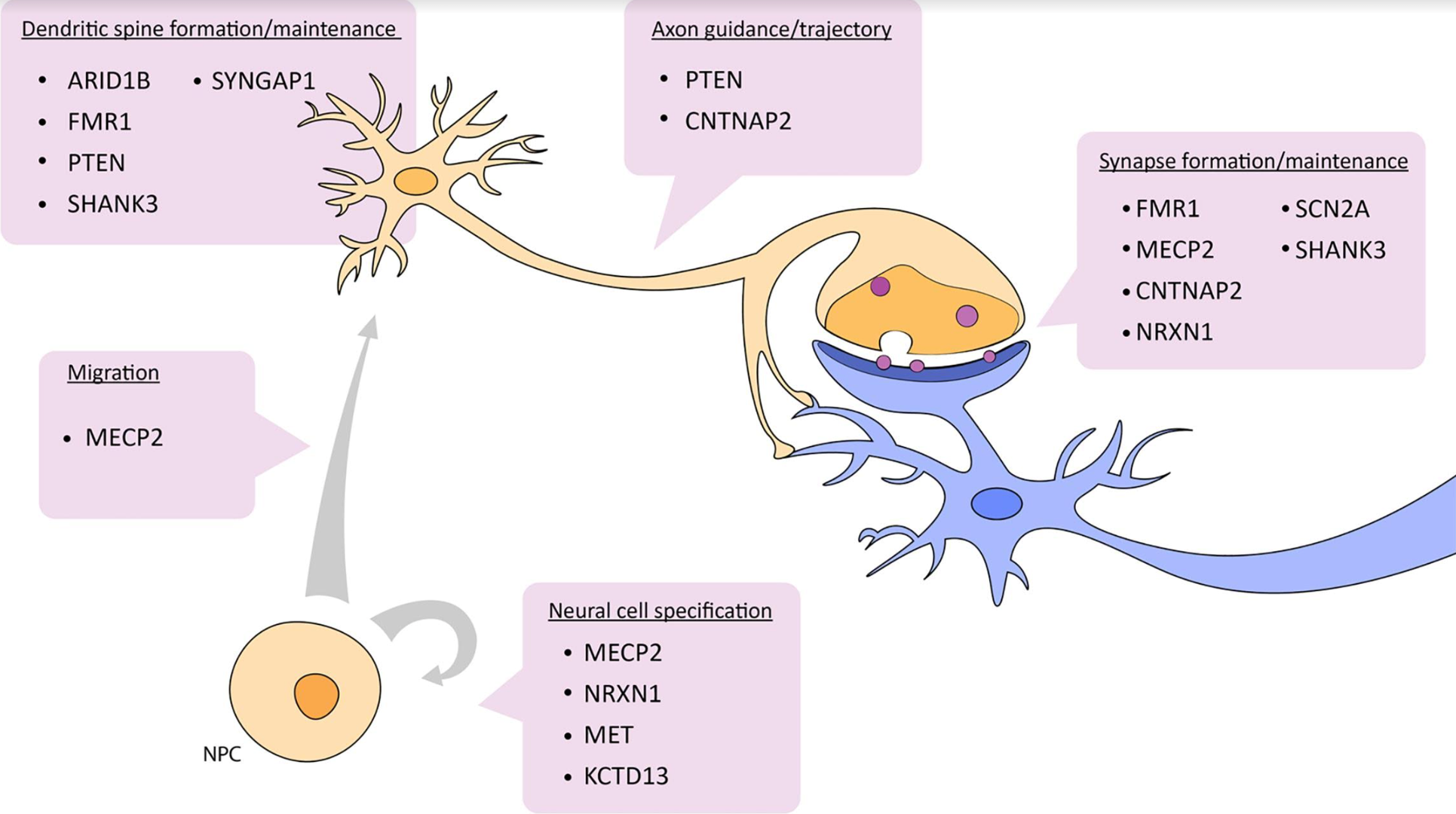
For example, we might want to know what genes are “turned on” (seen as a rise in specific mRNA levels) during cell migration. These investigators have found that methyl CpG binding protein 2 (MECP2) is increased in neuronal progenitors that are in the process of migration. (If you want to investigate any of the other genes listed, use your favorite search engine with the gene name plus “GeneCards” — e.g.
MECP2 GeneCards
— or simply search inside the GeneCards site.)
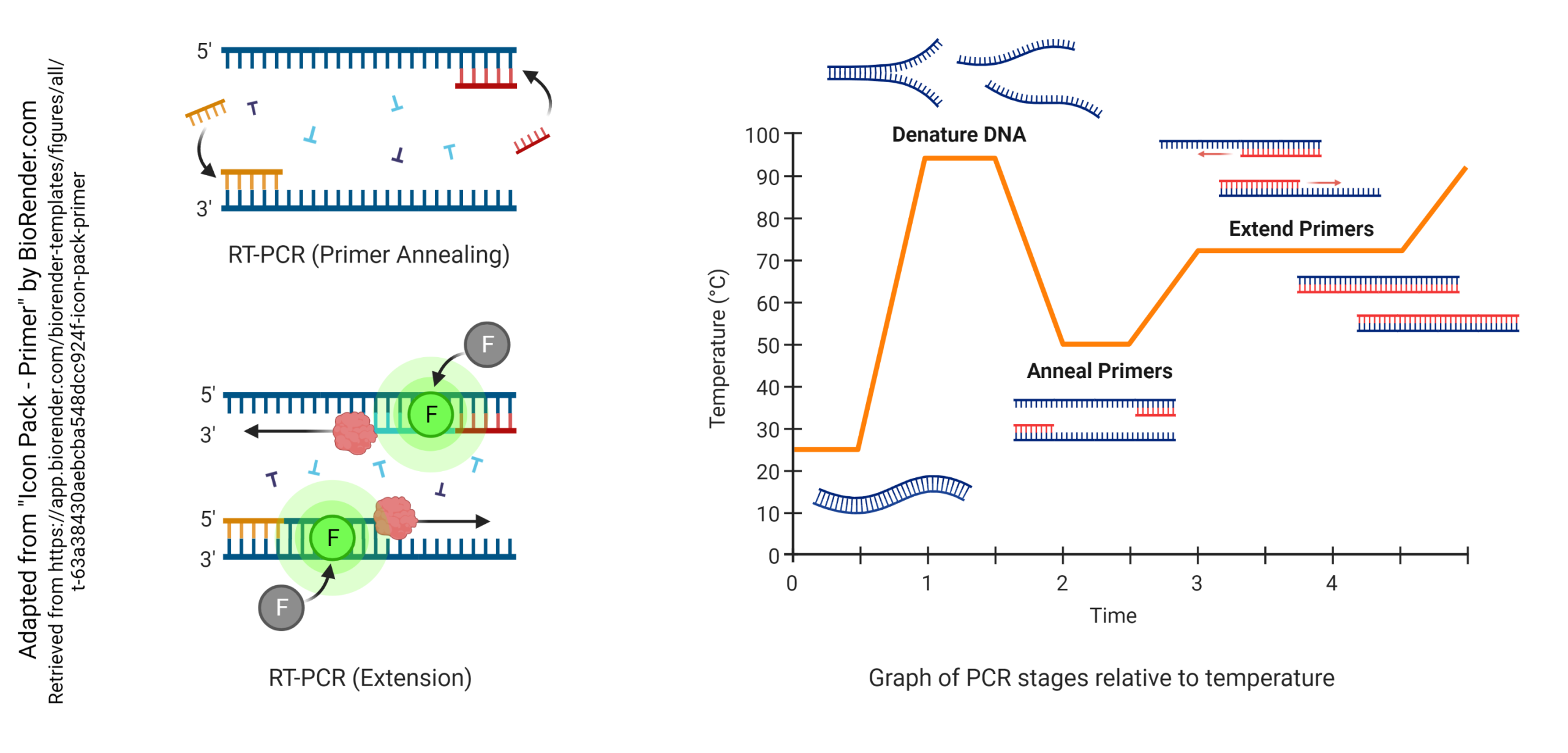 In the reverse transcriptase – polymerase chain reaction method, we carry out the following steps in order:
In the reverse transcriptase – polymerase chain reaction method, we carry out the following steps in order:
- total RNA is isolated
- the RNA which has poly(A) at the 3′ end is isolated from total RNA (this is presumed to be mRNA, since only mRNA carries a poly(A) tail
- the enzyme reverse transcriptase converts RNA to DNA, now called complementary DNA (cDNA)
In the polymerase chain reaction:
- the investigator has made two primers with an included fluorescent tracer (F), about 100 or so bases apart, which are complementary to opposite strands of DNA
- the DNA is denatured by heating (typically to 95°C)
- the primers are annealed to both strands of the DNA (typically at about 50°C)
- a thermostable DNA polymerase binds to the double-stranded region (where the primer is annealed) and extends the double-stranded sequence (typically at about 72°C)
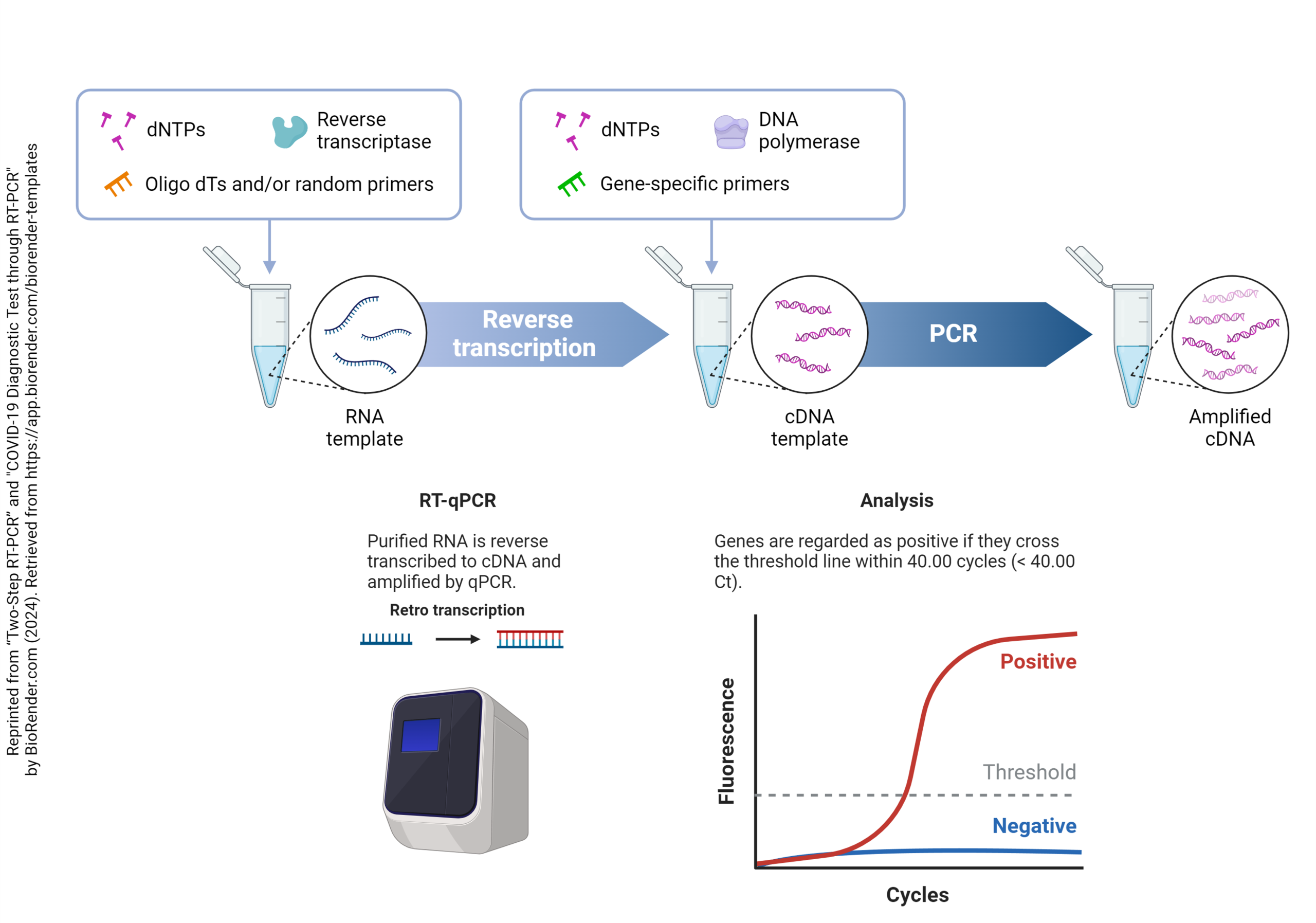 With each annealing cycle, the primers bind to each of the two strands of the double helix, facing off about 100 base pairs apart. Then, during the 72°C extension cycle, the thermostable DNA polymerase builds a new strand starting at the primers, each going 5′ to 3′ but in opposite directions. This builds two new strands with each cycle, which can then anneal to form twice as much double-stranded DNA as before the start of the cycle. One double-stranded DNA molecule becomes two, then four, then eight, and so forth. By the end of 30 cycles, each cDNA molecule is now 1,073,741,824 (230) DNA molecules. You can see that even a small handful of cDNA molecules in the original sample will be detectable after this many cycles. The cycle number where the threshold of detectability is crossed (Ct) depends on the concentration of cDNA in the original sample. A low Ct indicates a high initial concentration of that particular cDNA, while a high Ct indicates a low initial concentration. In this way, the amount of mRNA in the original sample can be determined relative to the concentration of a “reporter” or “housekeeping” gene.
With each annealing cycle, the primers bind to each of the two strands of the double helix, facing off about 100 base pairs apart. Then, during the 72°C extension cycle, the thermostable DNA polymerase builds a new strand starting at the primers, each going 5′ to 3′ but in opposite directions. This builds two new strands with each cycle, which can then anneal to form twice as much double-stranded DNA as before the start of the cycle. One double-stranded DNA molecule becomes two, then four, then eight, and so forth. By the end of 30 cycles, each cDNA molecule is now 1,073,741,824 (230) DNA molecules. You can see that even a small handful of cDNA molecules in the original sample will be detectable after this many cycles. The cycle number where the threshold of detectability is crossed (Ct) depends on the concentration of cDNA in the original sample. A low Ct indicates a high initial concentration of that particular cDNA, while a high Ct indicates a low initial concentration. In this way, the amount of mRNA in the original sample can be determined relative to the concentration of a “reporter” or “housekeeping” gene.
One significant limitation of RT-qPCR is that the investigator can only amplify one gene at a time. There is a need, in many experimental systems, to examine the amount of dozens or hundreds of mRNA species simultaneously. For example, as can be seen above, multiple genes interact in synaptic development. Many times one starts an experiment with no idea what those genes are. Or, in this example, a pathogen damages a cell, and we need to know which genes have been up-regulated, which down-regulated, and which are unchanged.
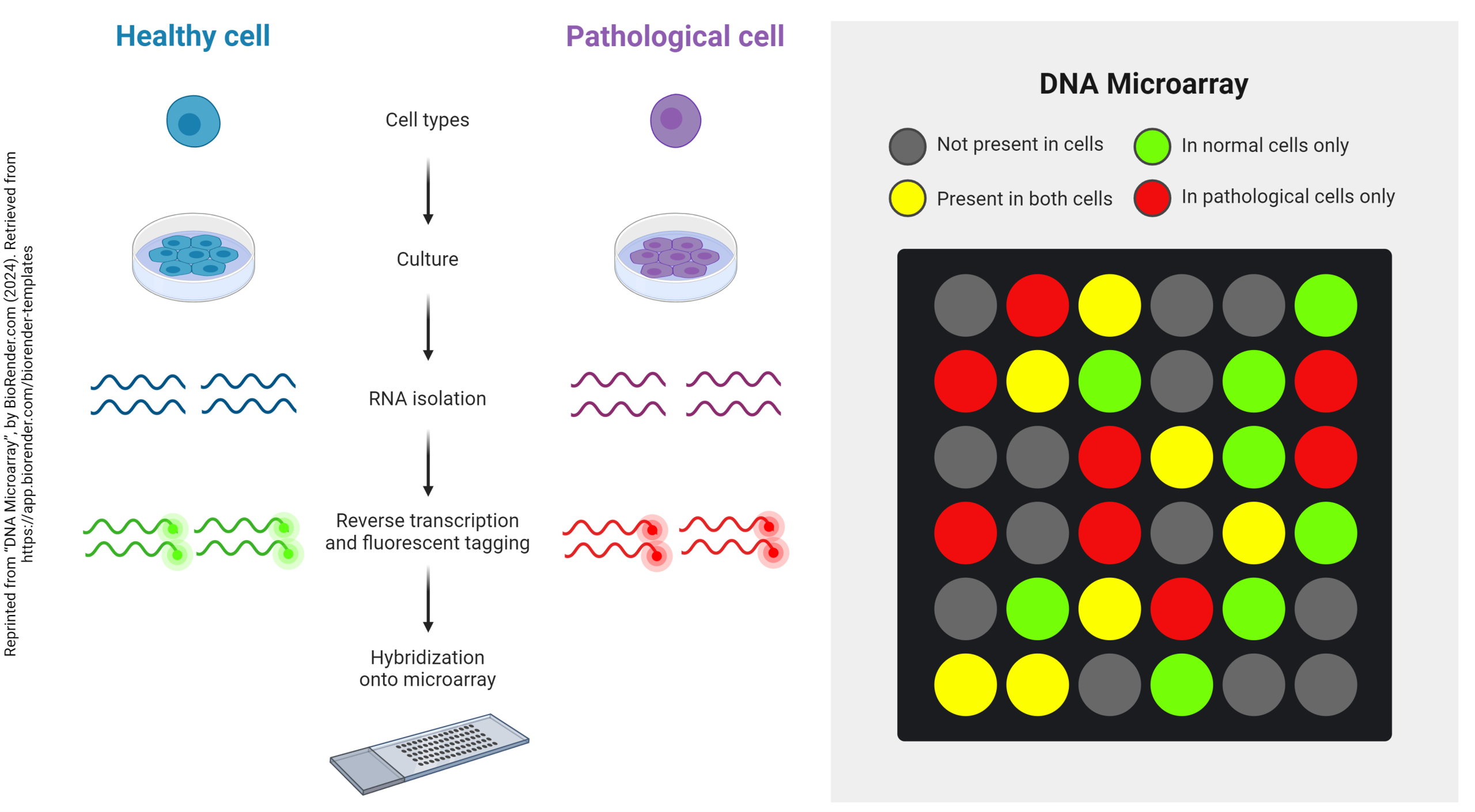 To help investigators understand what is happening in these circumstances, cDNA microarrays (gene arrays) were invented. This allows hundreds of genes (typically, 640) to be assayed simultaneously and compared between two different tissues — for example, two different developmental stages, such as before and after synaptogenesis, or two different states of health, such as normal and cancerous.
To help investigators understand what is happening in these circumstances, cDNA microarrays (gene arrays) were invented. This allows hundreds of genes (typically, 640) to be assayed simultaneously and compared between two different tissues — for example, two different developmental stages, such as before and after synaptogenesis, or two different states of health, such as normal and cancerous.
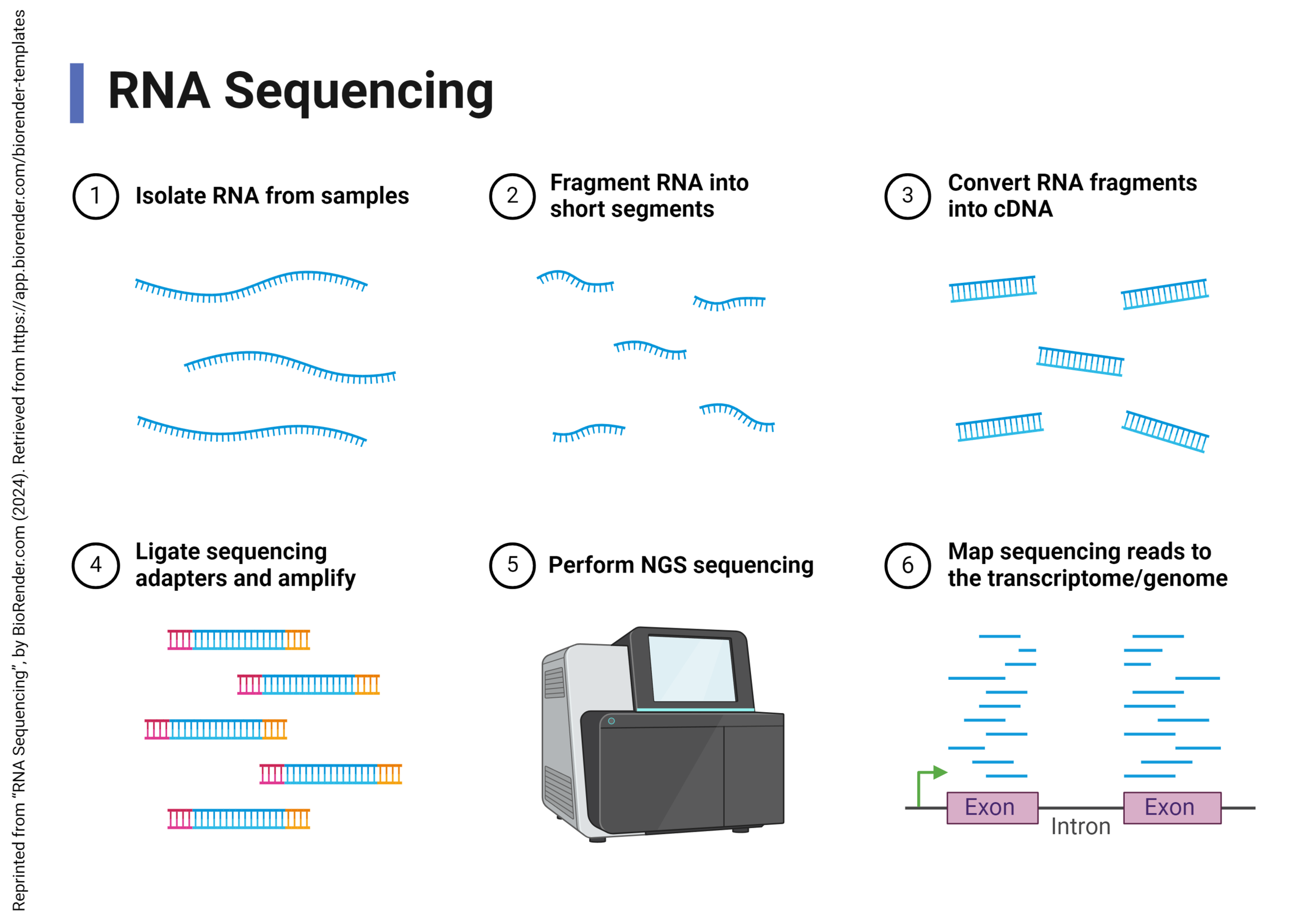 DNA microarrays have fallen out of favor and a much more cost-effective and accurate way to get the same information has been invented: RNA sequencing.
DNA microarrays have fallen out of favor and a much more cost-effective and accurate way to get the same information has been invented: RNA sequencing.
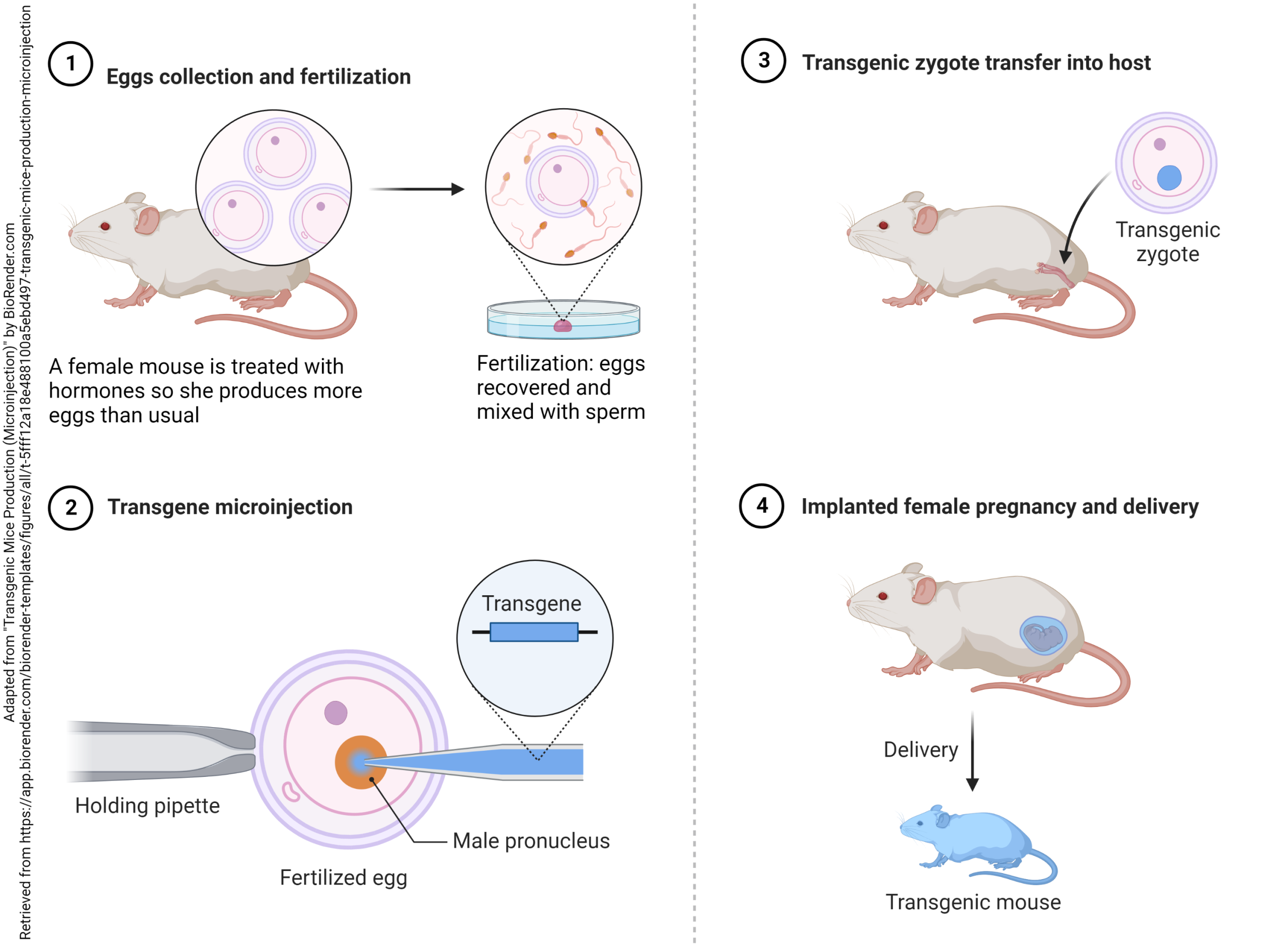 The 2007 Nobel Prize in Physiology or Medicine was awarded to Mario Capecchi, Martin Evans, and Oliver Smithies for the technique that came to be known as development of transgenic mice. Evans discovered the embryonic stem (ES) cell of the mouse embryo. This is the one cell in the early embryo that becomes 3,000,000,000 adult mouse cells or 10,000,000,000 adult human cells.
The 2007 Nobel Prize in Physiology or Medicine was awarded to Mario Capecchi, Martin Evans, and Oliver Smithies for the technique that came to be known as development of transgenic mice. Evans discovered the embryonic stem (ES) cell of the mouse embryo. This is the one cell in the early embryo that becomes 3,000,000,000 adult mouse cells or 10,000,000,000 adult human cells.
Novel DNA is introduced into ES cells by a technique called electroporation, and the modified ES cell can then be injected into a mouse embryo and implanted into a foster mother. The modified gene is now stable and can be transmitted to offspring in the usual way.
Two kinds of transgenic modifications are possible: knockout, where a normal gene is removed; and knock-in, where a novel gene is added.
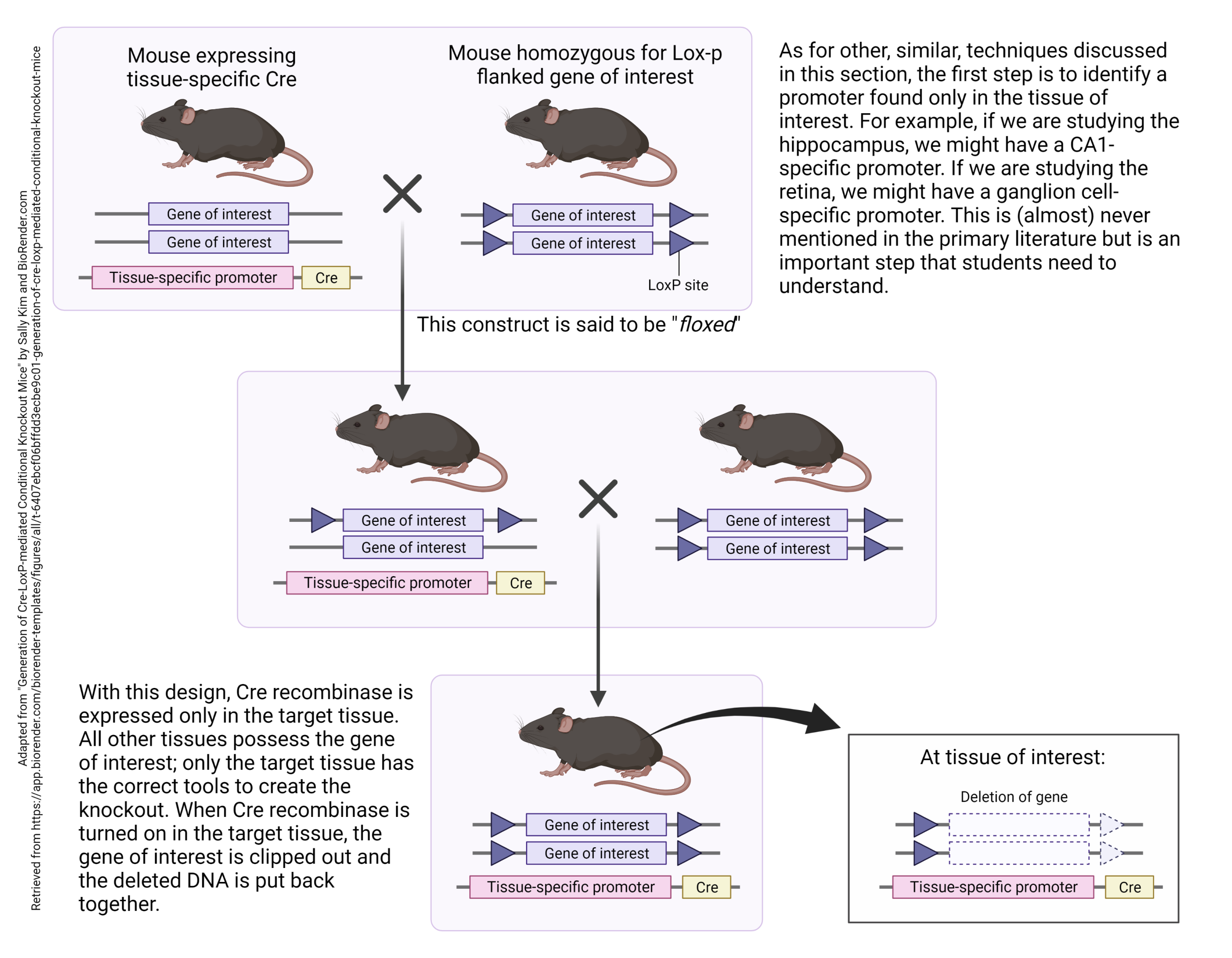 Often, knocking a gene in or out results in a dead animal. To avoid this, or to refine the location of the modified genes, one can build a tissue-specific promoter into the mouse genome just ahead of the knockout (or knock-in) gene.
Often, knocking a gene in or out results in a dead animal. To avoid this, or to refine the location of the modified genes, one can build a tissue-specific promoter into the mouse genome just ahead of the knockout (or knock-in) gene.
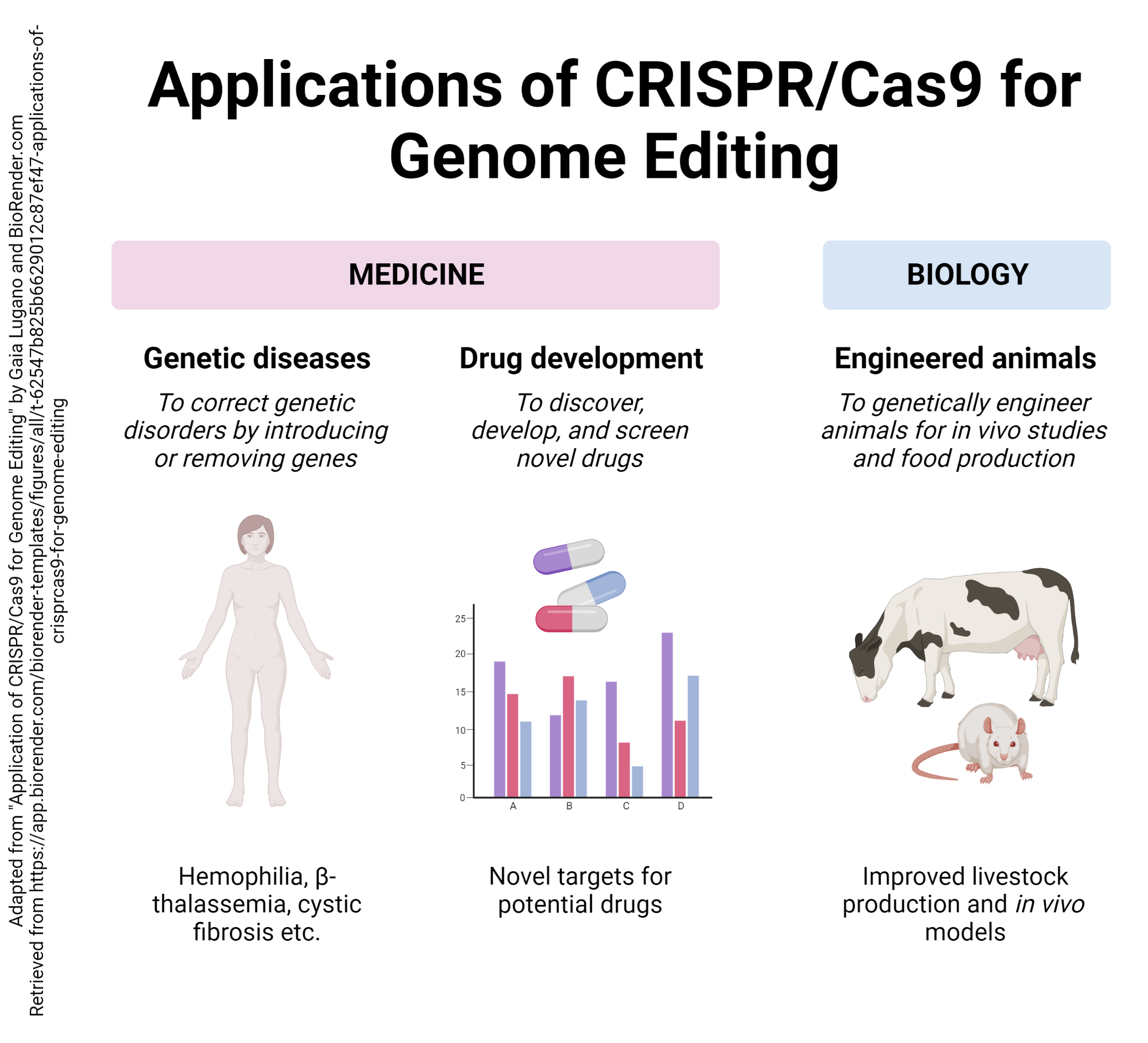 An even more targeted technique is based on a primitive defensive system used by bacteria, the CRISPR/Cas9 system.
An even more targeted technique is based on a primitive defensive system used by bacteria, the CRISPR/Cas9 system.
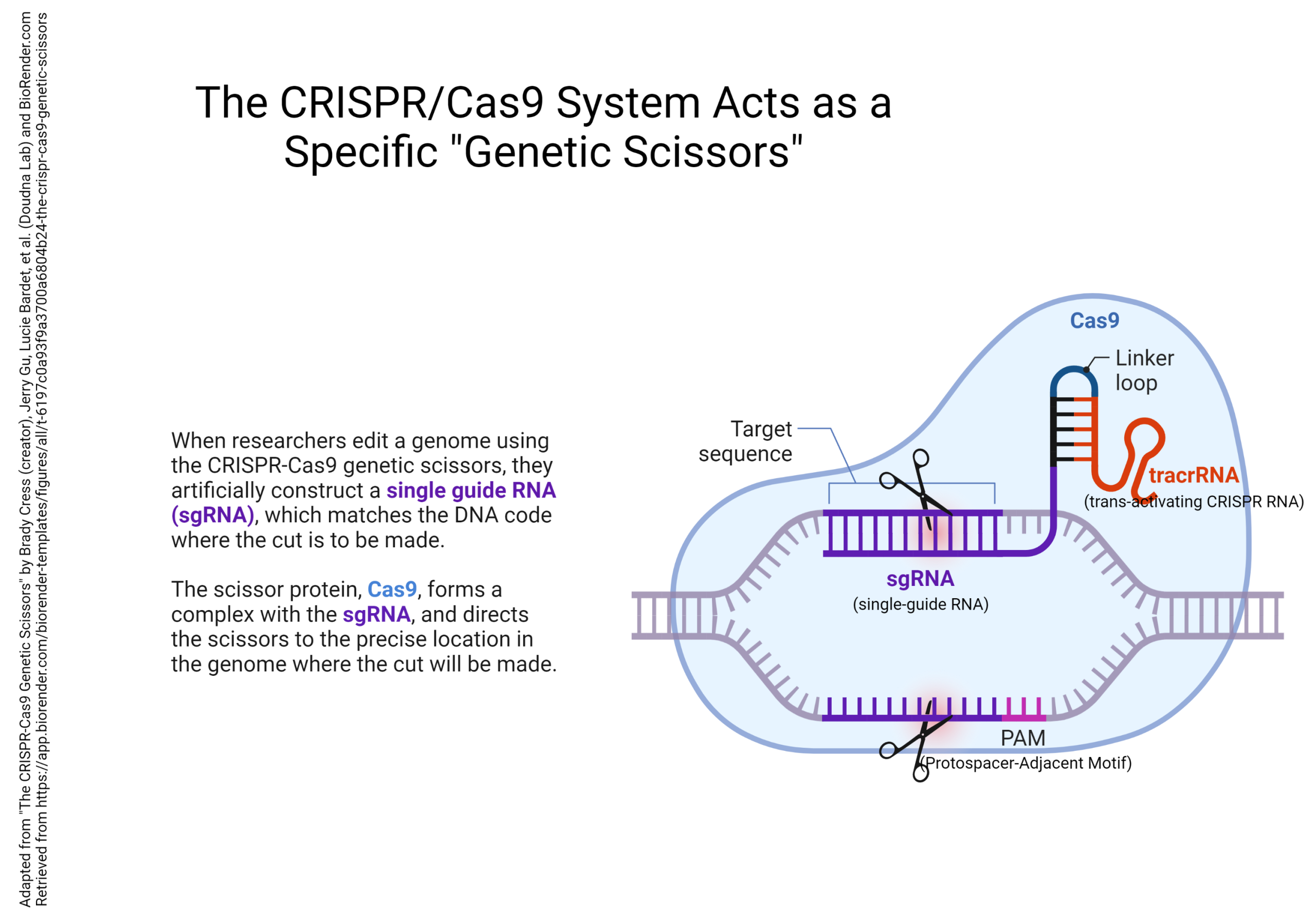
The gene sequence that is used to locate the cut is called a Clustered Regularly Interspaced Short Palindromic Repeat, or CRISPR, and the protein that does the cutting is called Cas9. The CRISPR/Cas9 system can be used to accurately edit specific gene sequences to whatever configuration the researcher wants.
Media Attributions
- Transcriptome in synapse formation © Victoria Rea and Terence J. Van Raay is licensed under a CC BY (Attribution) license
- Steps in RT-PCR © Wendy Jiang adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Quantitative rt-pcr © Wendy Jiang & Science Design adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Gene arrays © BioRender is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- RNA sequencing © Eunice Huang is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Production of Transgenic Mice by Microinjection © Catherine C adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Generation of conditional knockouts © Sally Kim adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Applications of CRISPR_Cas9 for Genome Editing © Gaia Lugano adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- CRISPR Uses a Cas9 Genetic Scissors © Brady Cress adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license

