John and Marcia Price College of Engineering
16 Good Predators and Where to Find Them
Vivian Marcoux and Jennifer Weidhaas
Faculty Mentor: Jennifer Weidhaas (Civil and Environmental Engineering, University of Utah)
Introduction
Wastewater surveillance is a valuable and frequently utilized tool in microbial ecology, with a history in epidemiology extending as far back as the 1854 Broad Street cholera epidemic (7). Communities of bacteria live in wastewater, forming ecological communities that interact with a variety of other organisms. Of note is the presence of predators, and if or how they interact with these bacterial communities. It is also valuable to search for the presence of antibiotic- resistance genes (ARG), due to their considerable scientific and medical implications. This investigation is therefore a survey of findings in wastewater in search of these predators.
Methods
Microscopy was performed at 4x, 10x, 20x, and 40x magnification via compound and inversion light microscope, documented via attached and labeled using attached Amscope MJ900 and Moticam BTW8 cameras and software. Slides were prepared, including unstained wet mount slides, and a variety of staining was performed: gram staining with crystal violet and safranin, methylene blue staining, malachite green staining with safranin. These slides were then inspected and findings cataloged. Samples were from wastewater sampled from the University of Utah campus and Nielsen Rehabilitation Hospital sewer system via grab and automatic capture. A fellow lab mate analyzed wastewater representative of that used in this survey for ARG via Cepheid Gene Xpert analysis.
Results
The invariable observation of this survey was a great quantity of bacterial colonies of varying size, predominantly arranged into flocs. This is typical of wastewater studies of this type. Bacteria was also present in filamentous colonies that mimicked fibers, which were also present, of organic and inorganic origin. Predators were visualized in this survey. This included apparent prokaryotes, as well as amoebaform and nematodous findings. This indicates that within the wastewater sampled, predators were present, and therefore likely to have interacted with bacterial colonies and impacted these ecological communities. The predators observed are typical in these types of studies(2,5), but this survey did not reveal the presence of several of the other potential varieties of predators.
Conclusion
This survey detected predators of prokaryotic, amoebaform, and nematodous nature in wastewater sampled from across the University of Utah, indicating that these ecological communities may include active predation that may impact bacterial population. Gene Xpert analysis by a labmate revealed the presence of multiple ARG in wastewater representative of that used in this study, which indicates a dangerous potential for the development of antibiotic resistance with larger health implication than those currently observed. These predators, as part of the ecological community in this wastewater, are part of the puzzle of bacterial survival, including antibiotic resistance. Further exploration of these predators has the potential to reveal potential interventions for this problem.
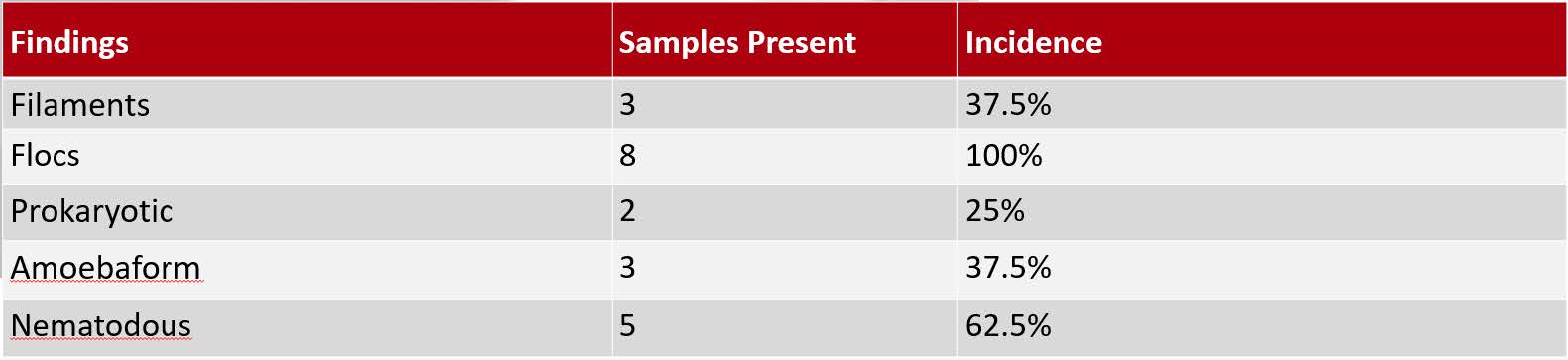
Table 1: Findings in Sampled Wastewater – Type and incidence of findings
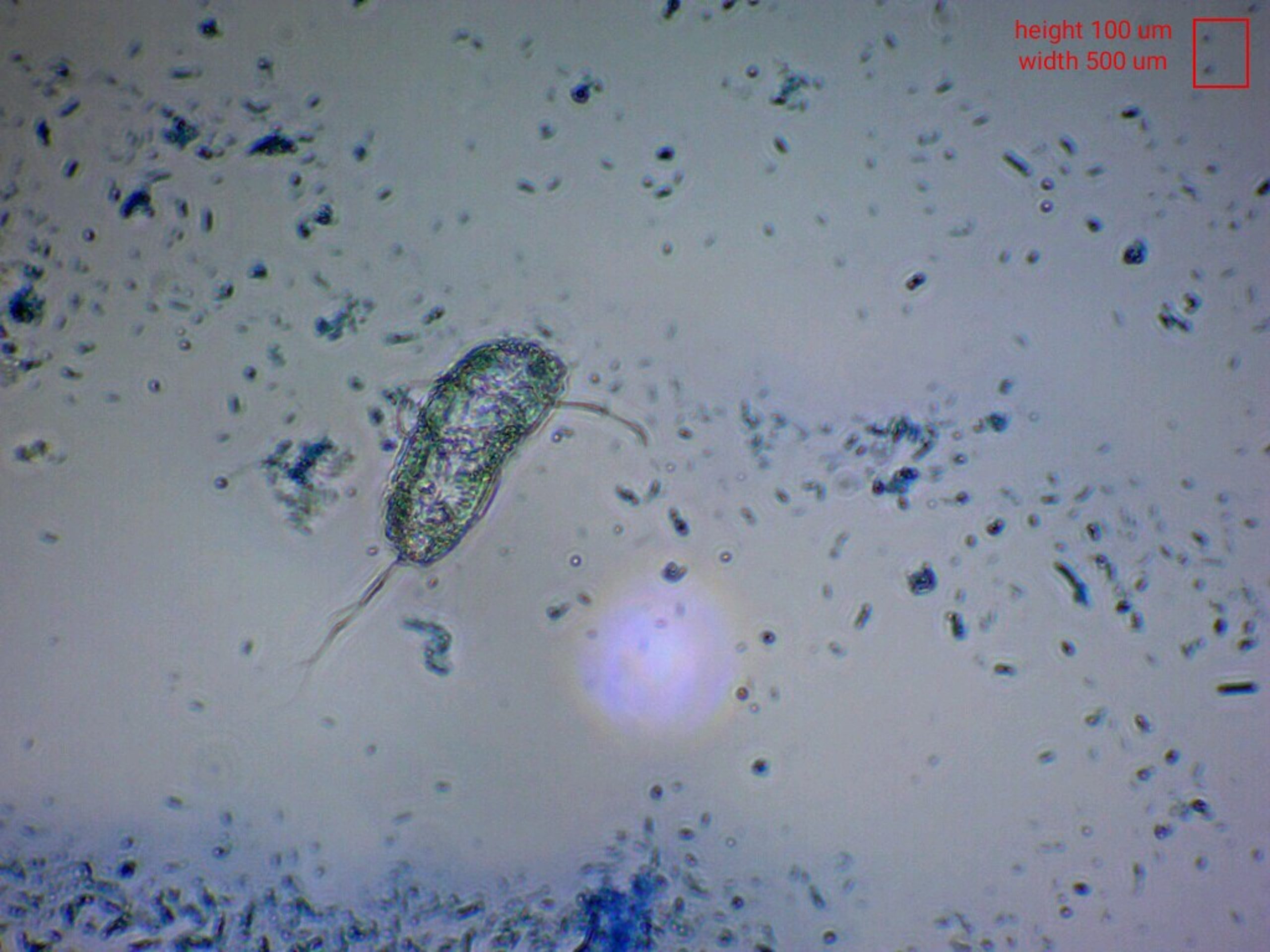 Figure 1. Flagellate visualized via methylene blue at 40X magnification brightfield inversion microscopy
Figure 1. Flagellate visualized via methylene blue at 40X magnification brightfield inversion microscopy
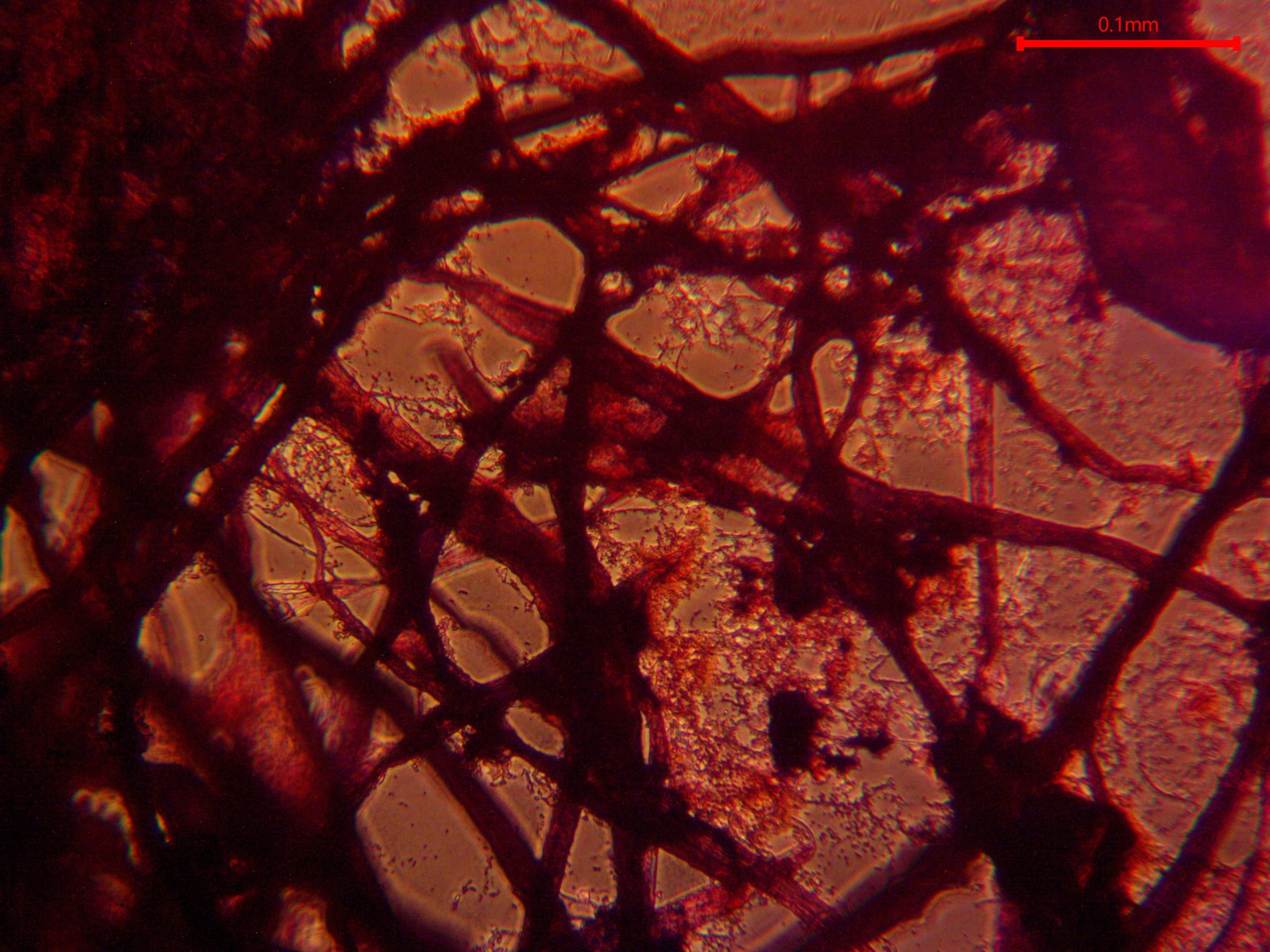
Figure 2. Filamentous bacteria visualized via gram staining at 20X magnification brightfield microscopy
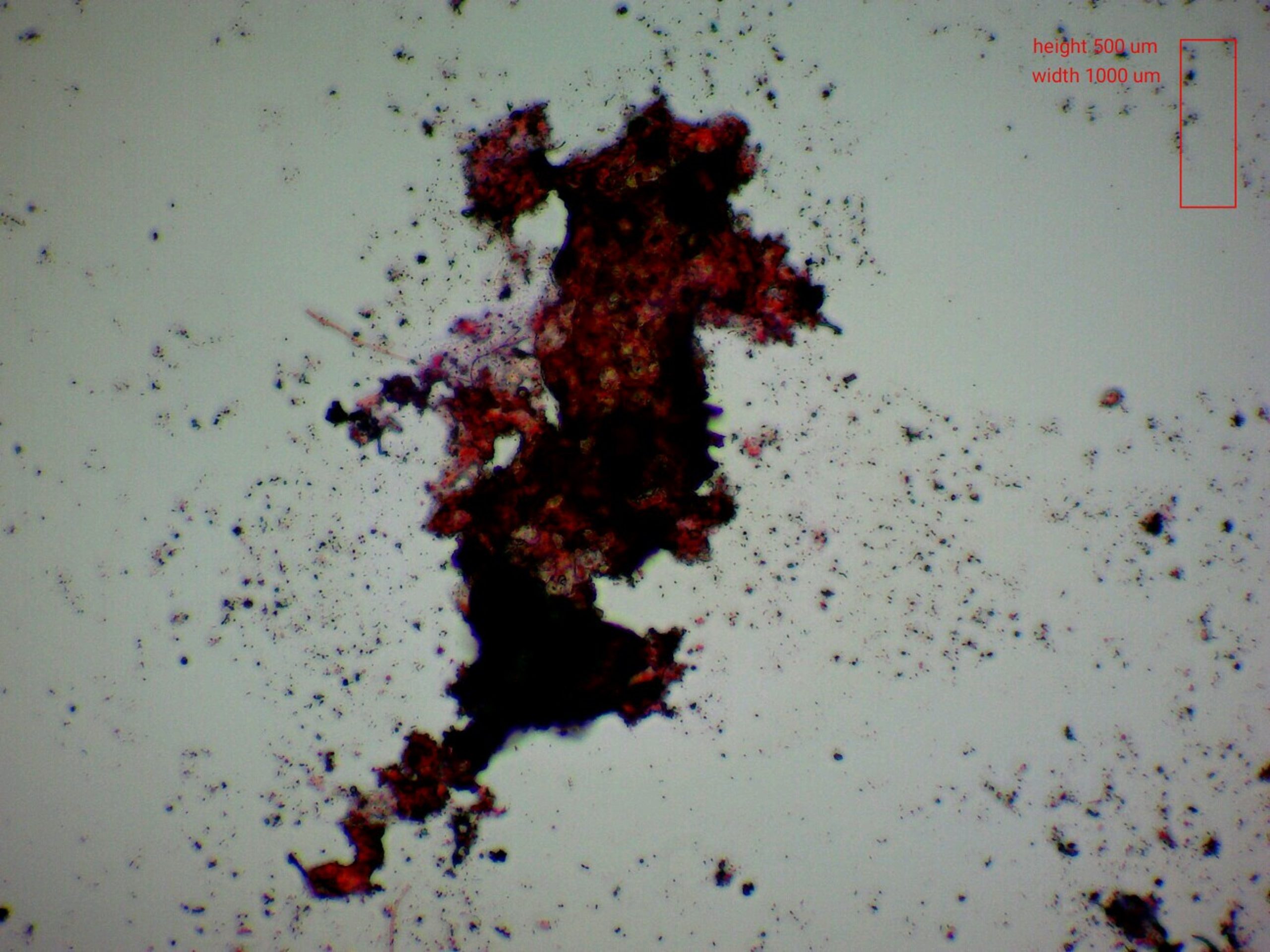
Figure 3. Bacteria floc, surrounded by smaller flocs and colonies, visualized via gram staining at 20X magnification brightfield microscopy
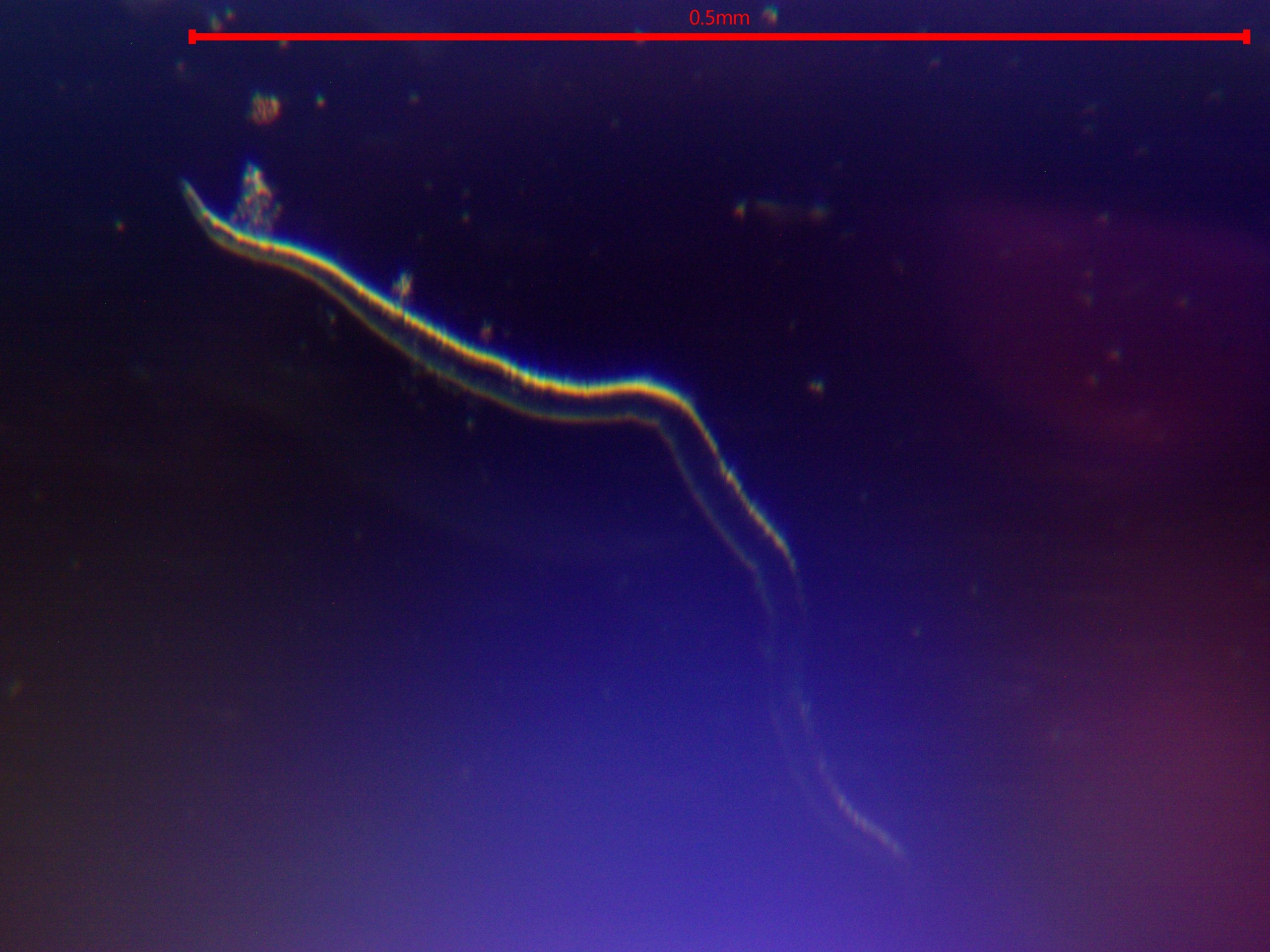 Figure 4. Nematodous finding visualized via unstained wetmount at 20X magnification darkfield microscopy.
Figure 4. Nematodous finding visualized via unstained wetmount at 20X magnification darkfield microscopy.
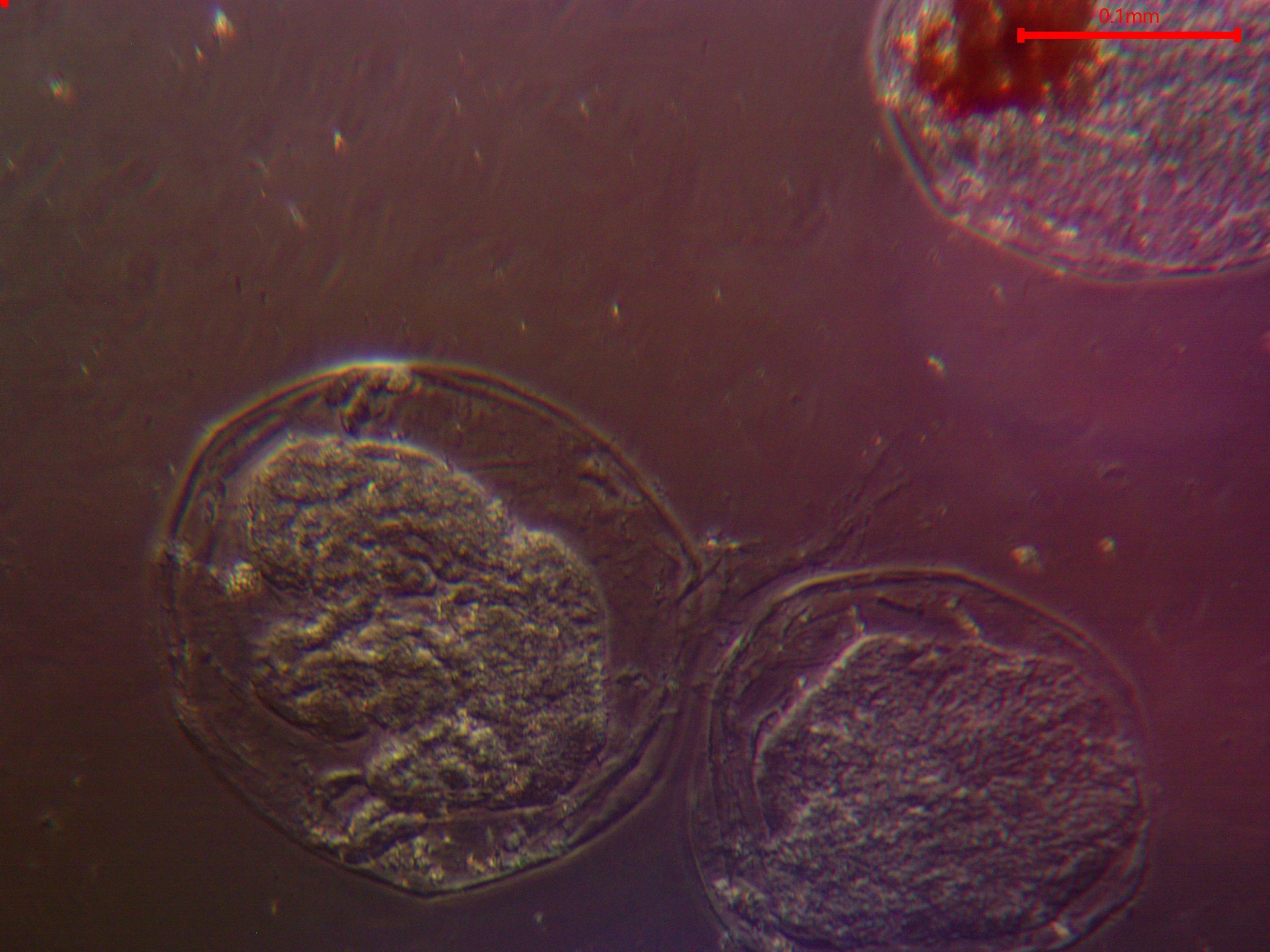 Figure 5. Amoebaform findings visualized via unstained wetmount at 20X magnification darkfield microscopy
Figure 5. Amoebaform findings visualized via unstained wetmount at 20X magnification darkfield microscopy
Footnote
- Burian, A., Pinn, D., Peralta-Maraver, I., Sweet, M., Mauvisseau, Q., Eyice, O., Bulling, M., Röthig, T., & Kratina, P. (2022). Predation increases multiple components of microbial diversity in activated sludge communities. The ISME Journal, 16(4), 1086–1094. https://doi.org/10.1038/s41396-021-01145-z
- Kaiser, G. E. (n.d.). Wastewater treatment microscopic organism identification guide [Wastewater Treatment Organism Identification]. Microscope World; MicroscopeWorld. https://www.microscopeworld.com/p-4415-wastewater-treatment-organism-identification.aspx
- Sims, N., & Kasprzyk-Hordern, B. (2020). Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environment International, 139, 105689. https://doi.org/10.1016/j.envint.2020.105689
- Singer, A. C., Thompson, J. R., Filho, C. R. M., Street, R., Li, X., Castiglioni, S., & Thomas, K. V. (2023). A world of wastewater-based epidemiology. Nature Water, 1(5), 408–415. https://doi.org/10.1038/s44221-023-00083-8
- van Dommelen, J. (n.d.). The microbiology of wastewater treatment—Life in the Aeration Tank: Bacteria, Protozoa, and Metazoa. Ohio EPA Compliance Assistance Unit. (Original work published 2020)
- Heck, N., Freudenthal, J., & Dumack, K. (2023). Microeukaryotic predators shape the wastewater microbiome. Water Research, 242, 120293. https://doi.org/10.1016/j.watres.2023.120293
- Tulchinsky, T. H. (2018). John snow, cholera, the broad street pump; waterborne diseases then and now. Case Studies in Public Health, 77–99. https://doi.org/10.1016/B978-0-12-804571-8.00017-2
- Zhang, L., Huang, X., Zhou, J., & Ju, F. (2023). Active predation, phylogenetic diversity, and global prevalence of myxobacteria in wastewater treatment plants. The ISME Journal, 17(5), 671–681. https://doi.org/10.1038/s41396-023-01378-0
Media Attributions
- 55776342_good_predators_and_where_to_find_them_-_marcoux
- flagellate
- fibers
- clumps
- nematode
- amoeba

