Spencer Fox Eccles School of Medicine
52 Investigating the Role of Tom70 in Mitochondrial-Derived Compartment Formation by Creating CRISPR/Cas9 Mutations in S. cerevisiae
Abby Wilson; Adam Hughes; and Zachary Wilson
Faculty Mentor: Adam Hughes (Biochemistry, University of Utah)
Abstract
Mitochondria are a vital organelle because they perform many essential metabolic processes and support energy production in cells. For example, mitochondria host biochemical reactions that support ATP production, amino acid metabolism, and the biosynthesis of lipids, heme, and iron sulfur clusters (Rutter and Hughes, 2015). Due to this central role in cell metabolism the loss of mitochondrial function can result in many age-associated metabolic and neurodegenerative disorders (Killackey et al., 2020). Our lab investigates pathways cells use to preserve mitochondrial homeostasis in response to metabolic and cellular stressors and as cells age. A main pathway our lab studies is mitochondrial-derived compartments (MDCs), a mitochondrial remodeling pathway that removes excess or damaged proteins from the outer mitochondrial membrane (OMM) (Hughes et al., 2016). Currently, it is unknown how MDCs form from the OMM or target cargo proteins into MDCs for degradation. We hypothesized that the OMM protein, Tom70, a protein involved in mitochondrial protein import, could be important for MDC formation because tom70∆ cells produce less MDCs. To examine how Tom70 supports MDC formation we utilized budding yeast, Saccharomyces cerevisiae, and CRISPR/Cas9 to mutate domains of Tom70. By studying a structural model of Tom70 and comparing sequence conservation alignments, we made amino acid substitutions in regions of Tom70 with high sequence conservation that were located within either the chaperone binding domain, precursor protein binding domain, or dimerization interface of Tom70. Subsequently, we analyzed the effect of these mutations on supporting MDC formation and cell proliferation. Our preliminary results suggest that utilizing the CRISPR/Cas9 systems may be a beneficial method to identify functional residues within Tom70 that support MDC formation.
Methods
To identify regions in Tom70 that could be important for its function we analyzed a protein sequence alignment of Tom70 homologs from several different yeast species. We predicted that important functional amino acids in Tom70 would be highly conserved across species. We then compared conserved regions to a structural model of Tom70 (Wu and Sha, 2006) and of Tom71, a paralog to Tom70 (Li et al., 2009) to compile a list of different mutations within the chaperone binding region, preprotein binding pocket, and dimerization interface of Tom70 that might be required for Tom70 function.
We substituted groups of 3-5 amino acids in Tom70 for alanines using a CRISPR/Cas9-based technique to modify the TOM70 gene in the yeast genome (Guo et al., 2018). These mutations were constructed in a yeast strain that lacked Tom71 (tom71∆) and also expressed the mitochondrial proteins Tcd2-GFP and Tim50-mCh (which helped with MDC identification). Within the tom71∆ Tcd2-GFP Tim50-mCh strain we integrated CAS9 and subsequently transformed these cells with plasmids containing CRISPR guide RNAs and a template DNA sequence that could be used for homology-directed repair to create targeted mutations in the TOM70 gene.
After transforming cells with the plasmids that contained the guide RNAs, we isolated colonies, obtained genomic DNA, amplified the TOM70 gene, and sequenced the gene to confirm they had the substitutions we designed. Finally, we performed growth assays and MDC assays on the Tom70 mutants to test for temperature-sensitive growth phenotypes and changes in quantity of MDC formation. For MDC assays, cells were treated with DMSO (control) or Rapamycin (Rap), an inducer of MDCs. After treatment for two hours, live yeast cells were imaged on a widefield fluorescence microscope, and MDCs were quantified.
Results
We obtained results for eight different mutants containing the following mutations in Tom70: L207L211, L371-M373, P433-L437, Y442-E444, L564-E567, E577-Q580, T583-E586, which were all substituted to alanine and a 19 amino acid deletion, ∆226-244. All these mutations were confirmed with sequencing of the whole Tom70 gene, and some contained off-site mutations. With each of these mutants we performed growth assays growing them at 30°C and 38°C on SD-complete plates to test if the mutations caused a temperature-sensitive growth phenotype (Figure 1). MDC assays were also performed for each strain after being treated with DMSO and Rap. Percentage of MDCs under both conditions for each strain was recorded (Figure 2). Representative images are shown for some mutants where MDCs are indicated with a white arrow (Figure 3).
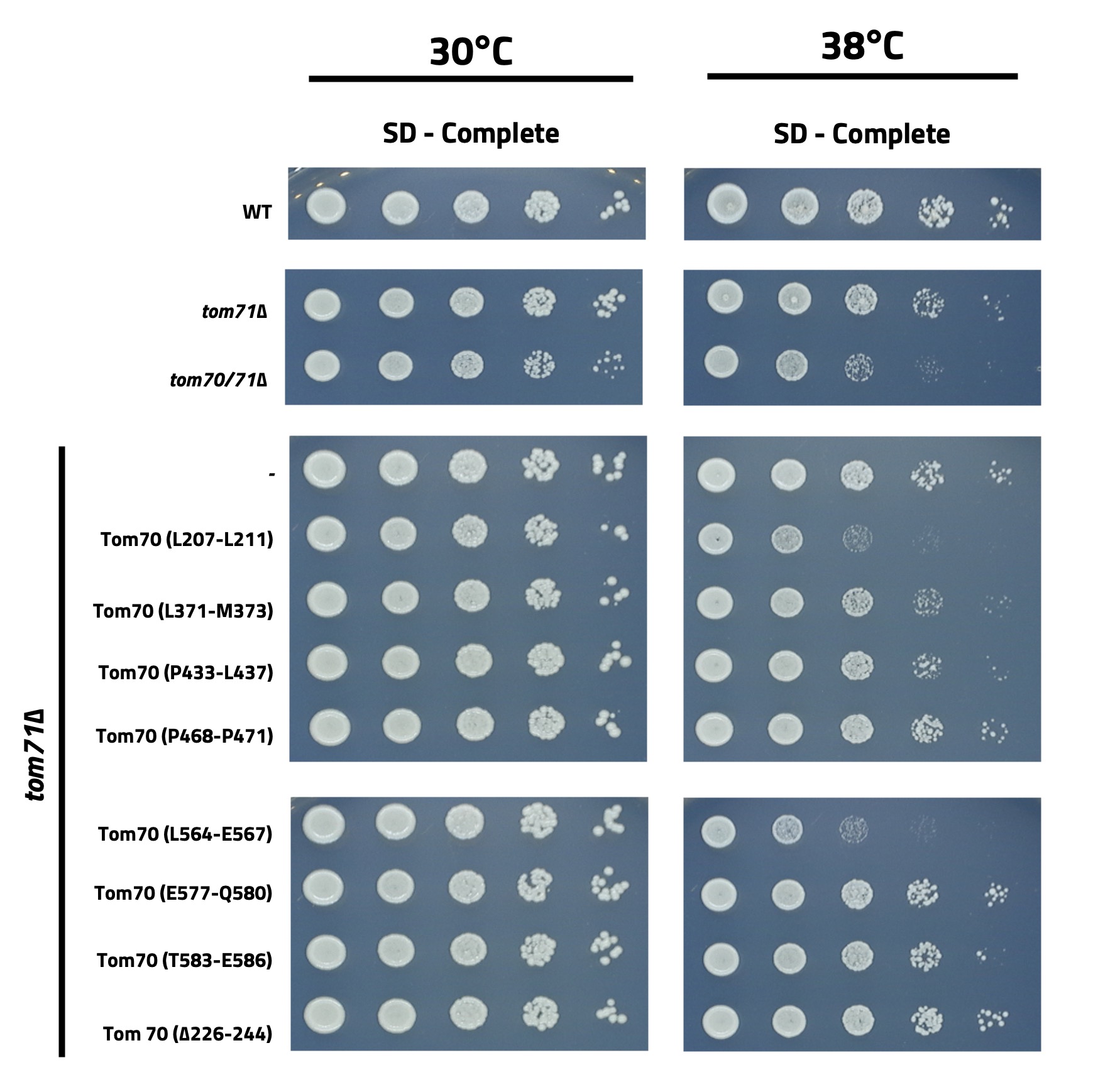
Figure 1. Images of a growth assay at two different temperatures of the yeast strains indicated.
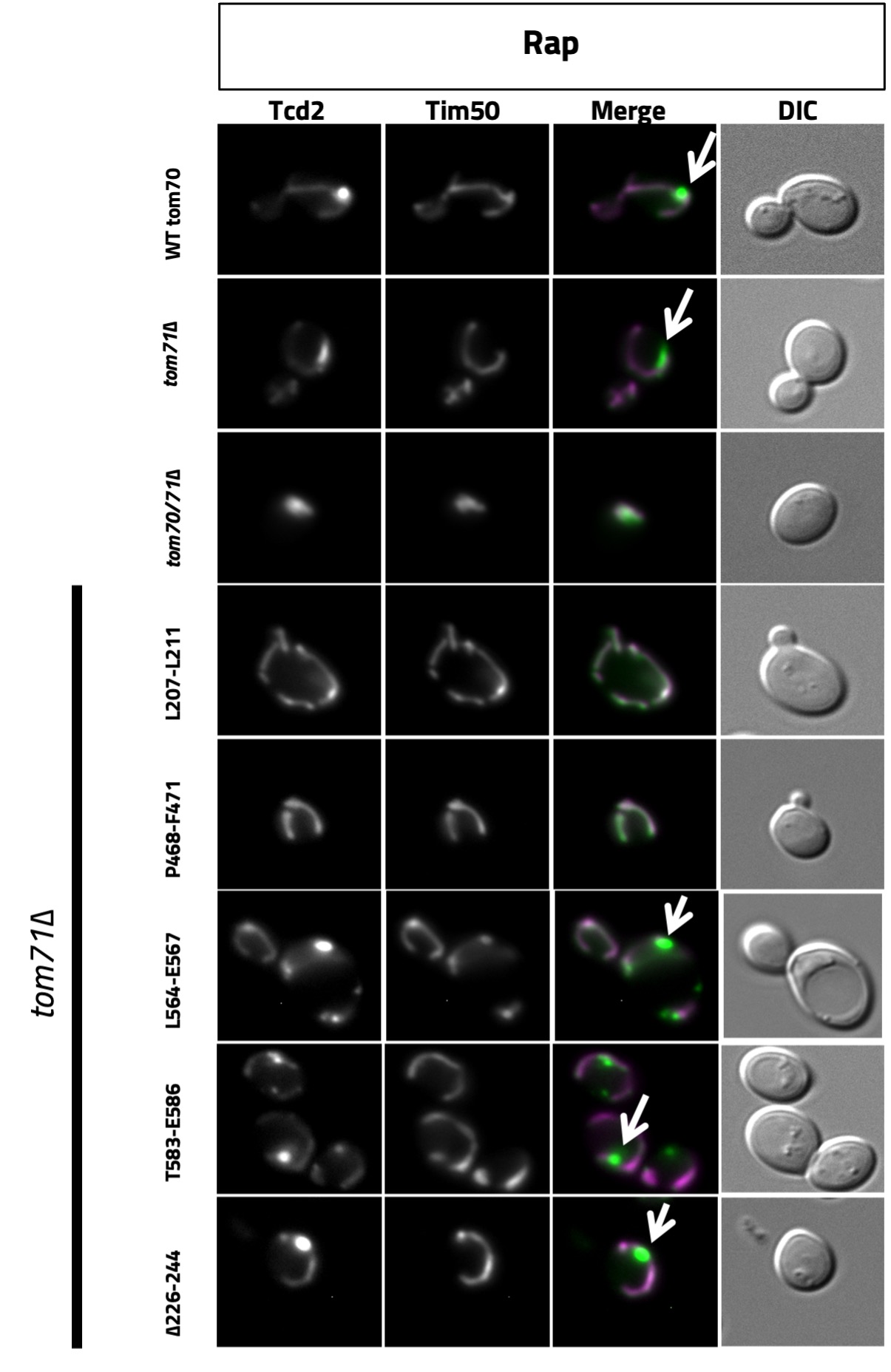
Figure 2. Representative widefield fluorescence microscopy images for the yeast strains indicated, all of which expressed Tcd2-GFP and Tim50-mCh. In the merged images Tcd2 can be seen in green while Tim50 can be observed in magenta. MDCs that were observed are indicated with a white arrow.
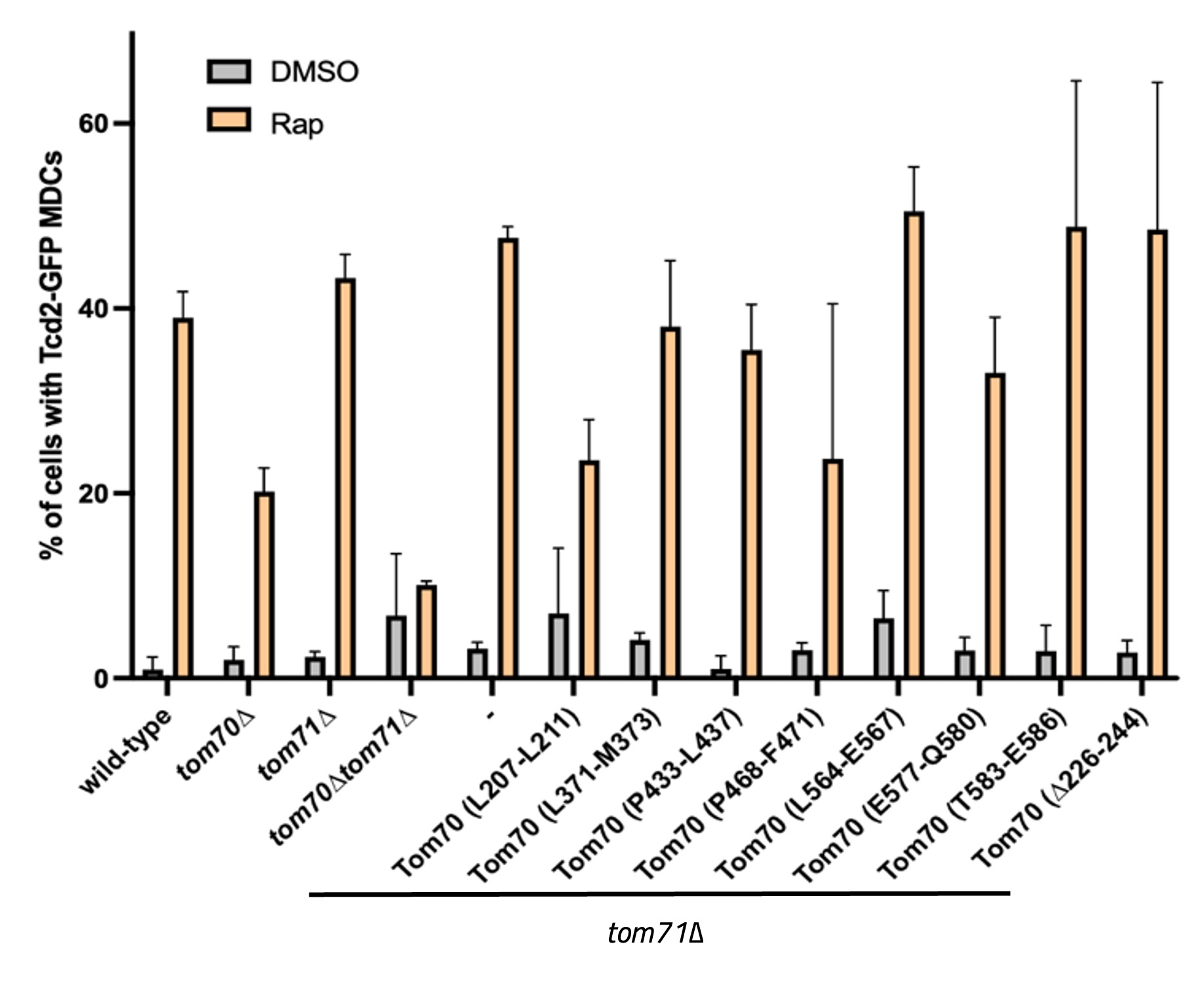
Figure 3. Graph of the percent of MDCs formed in the yeast strains indicated observed after a two-hour treatment of DMSO or Rapamycin (Rap). Related to the experiment shown in Figure 2.
Discussion
The temperature-sensitive growth phenotype recognized in the two mutant strains (L207-L211 and L564-E567) indicate that these mutations impair Tom70 function potentially by disrupting Tom70 from chaperoning proteins during heat stress. Interestingly, these two mutations land in separate domains of Tom70, where L207-L211 is a mutation in the dimerization interface of the protein while L564-E567 is a highly conserved region of Tom70 near the pre-protein binding pocket (Wu and Sha, 2006). However, we noticed the L207-L211 mutant contained additional off-site mutations and it would be worthwhile to examine if these mutations alter the stability of the Tom70 protein, via a Western blot assay, as this could also explain the results we observed. As for the strains producing less MDCs, the P468-F471 mutation in Tom70 is intriguing as it is in the pre-protein binding pocket of Tom70, and while this mutant strain produced less MDCs it did not have a temperature-sensitive growth defect. These results could implicate that these residues are involved in Tom70’s ability to help the cell produce MDCs.
Overall, our results began to identify unique regions within Tom70 that were important for different functions of Tom70 in relation to supporting cell growth at high temperature or supporting MDC formation. Due to the promising nature of these results, it would be beneficial to move forward with this method to continue screening regions of Tom70 for functional differences. Results like these can help scientists better understand the functional role of different regions of the protein. This can be done using the CRISPR/Cas9 method utilized here or by designing different plasmids with the alanine substitutions.
Conclusion
In our results we noticed a temperature-sensitive growth phenotype in the strains with Tom70 mutations at L207-L211 and L564-E567 that matched the growth phenotype of the tom70/71∆ strain. We also noticed lower MDC counts in the strains with Tom70 mutations at L207-L211 and P468-F471. A structural representation of Tom70 colored with the three mutations causing phenotypic differences can also be observed (Figure 4).
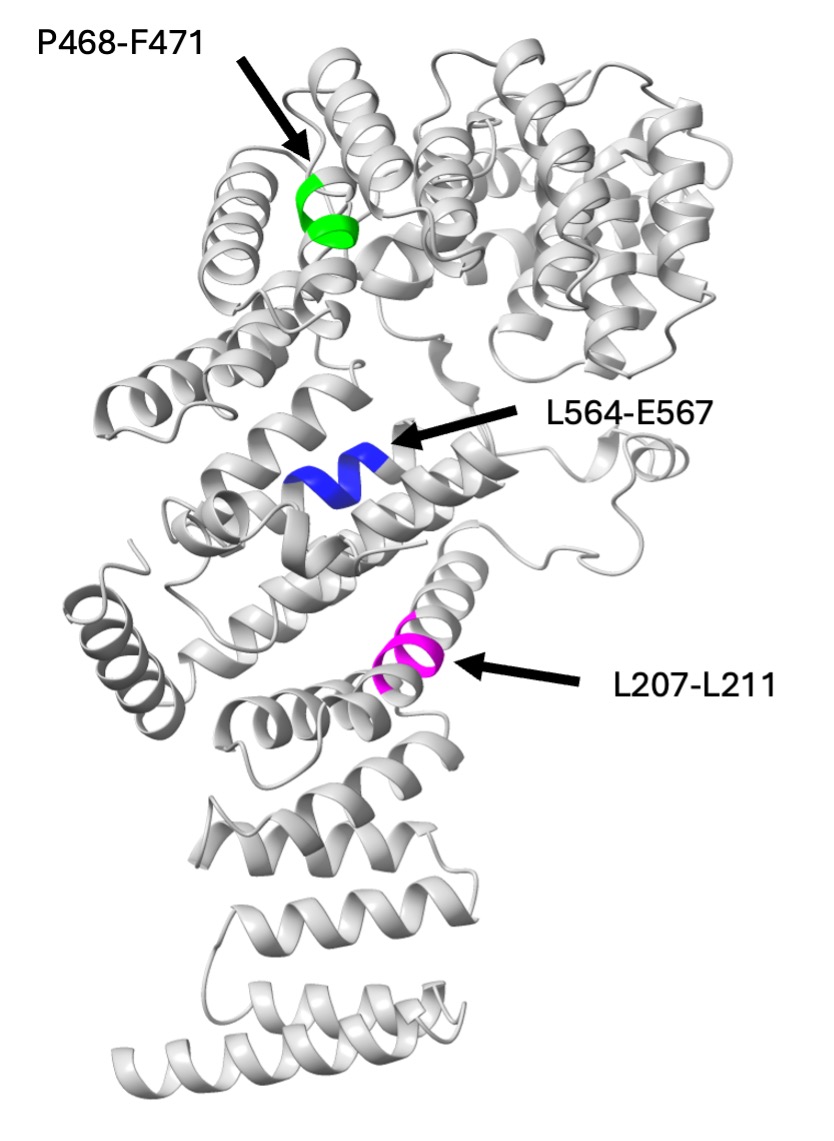
Figure 4. Structure of Tom70 highlighting the three mutations causing either a decreased MDC formation or temperaturesensitive growth phenotype. All are highly conserved regions, but L207-L211 is on the dimerization interface, P468-F471 in the preprotein binding pocket, and L564-E567 is near the preprotein binding pocket.
Bibliography
Guo, X., Chavez, A., Tung, A., Chan, Y., Kaas, C., Yin, Y., Cecchi, R., Garnier, S., Kelsic, E., Schubert, M., DiCarlo, J., Collins, J., Church, G. 2018. High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR–Cas9 in yeast. Nat Biotechnol 36: 540–546. doi: 10.1038/nbt.4147
Hughes AL, Hughes CE, Henderson KA, Yazvenko N, Gottschling DE. (2016). Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife 5:e13943. doi: 10.7554/eLife.13943
Killackey, S.A., Philpott, D.J. and Girardin, S.E. (2020). Mitophagy pathways in health and disease. Journal of Cell Biology, 219(11). doi: 10.1083/jcb.2020004029
Li J, Qian X, Hu J, and Sha B. (2009). Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem 284(35):23852-9. doi: 10.1074/jbc.M109.023986
Rutter J, Hughes AL. (2015). Power2: The power of yeast genetics applied to the powerhouse of the cell. Trends in Endocrinology & Metabolism 26:59–68. doi: 10.1016/j.tem.2014.12.002
Wu Y, Sha B. (2006). Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nature Structural & Molecular Biology 13, 589-593. doi: 10.1038/nsmb1106
Media Attributions
- 137489262_figure_1
- 146879582_figure_2
- 146879585_figure3
- 146881378_figure_4(1)

