College of Science
64 Investigating the Role of the Protein Interactions Domains of UBR5 in Mantle Cell Lymphoma
Cassandra Burdick; Shannon Buckley; and Deborah Kajewole
Faculty Mentor: Shannon Buckley (School of Biological Sciences, University of Utah)
Introduction
Mantle cell lymphoma (MCL) is a rare and aggressive non-Hodgkin’s lymphoma. ¹ Unfortunately, limited therapies for MCL are currently available suggesting a need to further unravel molecular mechanisms regulating transformation and progression of the disease. ² The ubiquitin E3 ligase UBR5 is mutated in ~18% of MCL patients with the majority of mutations in the HECT domain. ² E3 ubiquitin ligases serve as the substrate-recognizing component for protein degradation by the ubiquitin proteasome system. ² Our mouse model mimics patient mutations that leads to defects in B cell activation and germinal center formation, and proteomic studies reveal up-regulation of proteins associated with mRNA splicing via the spliceosome in B cells lacking the HECT domain of UBR5. ² The MLLE domain of UBR5 remains at the C-terminus, and MLLE domains have been associated with RNA processing, including splicing.³ Using the information, we will form a better understanding of the role of UBR5 in MCL with the goal of providing insights to mantle cell lymphoma transformation, progression and potential future therapeutics targets. To decipher the role of the MLLE and HECT domains in MCL, we knocked out the MLLE and HECT domains in MCL cell lines, JEKO1 cells by CRISPR/Cas9. Utilizing Nucleic gel techniques, we will define domain interactions and protein abundance changes specific to the MLLE and HECT domains through heterozygous and homozygous knockouts.
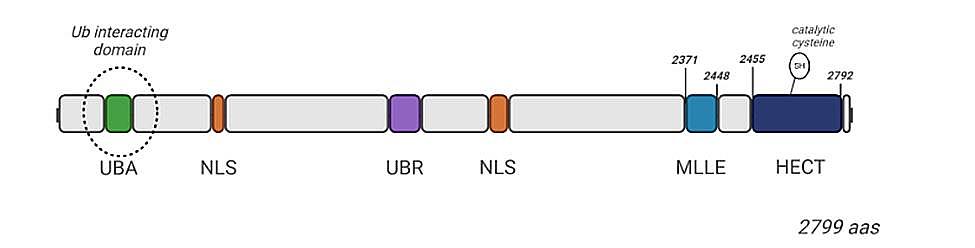
Figure 1: Kajewole, Dr. Deborah. (2024) [MLLE and HECT domains]. Biorender
Materials and Methods
Cell Lines Culture and CRISPR Preparation
JEKO1 MCL cell lines were cultured in RPMI-1640 medium supplemented with 0% fetal bovine serum and 1% penicillin-streptomycin at a 37°C incubation until 50-70% confluent. Cells were counted using a hemocytometer to ensure a concentration of 2x 10^5 to 2 x 10^6 cellars per 100 µ reaction. The collected cells were then washed with 5 mL of sterile 1x PBs, centrifuged, and removed from the media supernatant.
Formation of the Ribonucleoprotein complex
3 µL of JEKO1 MCL cells were resuspended in Buffer R (Neon Transfection kit), 4.5 µL Cas9 protein, and 7.5 µL the single guide RNA (sgRNA.) RP complex was left at room temperature for 5-10 minutes for incubation.
Thermo Fisher Neon Transfection
A Neon tube was filled with 3 mL of Electrolytic Buffer E2 and inserted into the pipette station. Neon Pipette tips were attached to the Neon Pipette to collect 100 µL of the RNP/Cell mixture
and interested in the mixture and inserted into the pipette station. The Neon Transfection system was set to electroporate at 1700V, 20ms after start was pressed.
Preparation for testing
The electroporated cells for the MLLE and HECT domain knockout attempts were transferred into pre-warmed media in a 6-well plate for a 24-72 resting period before being seeded into a 96-well U-bottom plate. After one week of growth, the electroporated wells were transferred into four 24-well U-bottom plates with 1 mL of medium and 500 µL of cells in each well.
Testing
gDNA was extracted from each of the 4 24-well plates and combined in a mastermix. The HECT master mix was made of a combination of 10 µL of Taq Polymerase, 7 µL of H2O, HECT forward Primer (F-intron 57- AAGCAGCTTCAGCCAAATTA), HECT reverse primer (R-intron 58- AGCTACTCTCTTCCTAGTAGTATGC), and 2 µL of DNA from the HECT knockout cells. The MLLE master mix was made of a combination of 10 µL of Thermo Fisher Phusion master mix, 7 µL of H2O, MLLE forward Primer (F-intron 49- CATGCCTGTAATTTCCCAGCTACT), MLLE reverse primer (R-intron 51- CTTGTCACCTGGAAAAAGG), and 2 µL of DNA from the MLLE knockout cells. These were then run in the thermocycler and injected into a Nucleic Acid gel to be visualized.
Results

For HECT, clone 1.8 had the heterozygous knockout, indicated by both having the wildtype band at around 4,000 bp and the knockout band at around 1,000 bp. The goal of finding heterozygous (both the knockout and WT band) indicates that the knockout was most likely successful in the HECT domain. Through confirmation and finding the homozygous band, the knockout can be deemed successful.
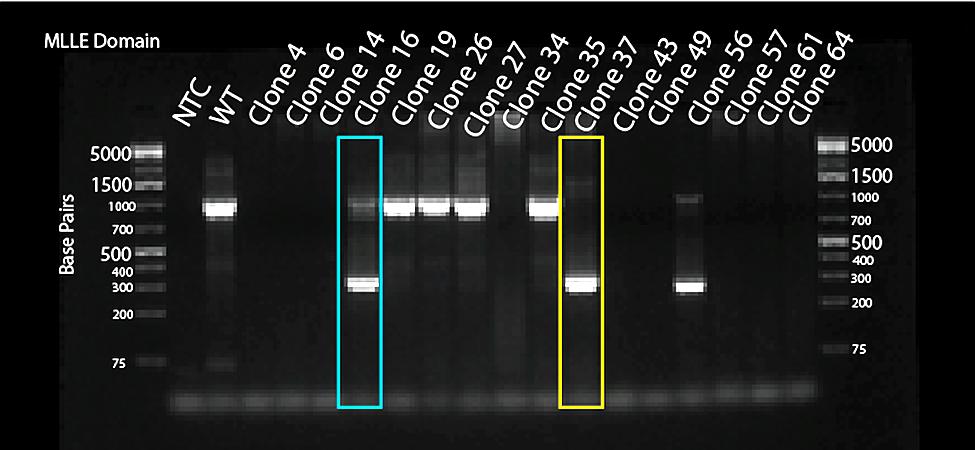
For MLLE, clones 16 and 56 had the Heterozygous knockout, indicated by both having the wildtype band at around 1,000 bp and the knockout band at around 300 bp. Also, for MLLE, clone 37 had the Homologous knockout, indicated by only having the knockout band at 300 bp. The goal of finding the homologous (just the knockout band) and heterozygous (both the knockout and WT band) was accomplished in the MLLE domain, indicating that the knockout was successful.
Bibliography
- National Cancer Institute. (n.d.). Mantle cell lymphoma treatment (PDQ®)–patient version. National Institutes of Health. Retrieved from https://www.cancer.gov/types/lymphoma/patient/mantle-cell-treatment-pdq
- Swenson, S. A., Gilbreath, T. J., Vahle, H., Hynes-Smith, R. W., Graham, J. H., Law, H. C.-H., Amador, C., Woods, N. T., Green, M. R., & Buckley, S. M. (2020). UBR5 HECT domain mutations identified in mantle cell lymphoma control maturation of B cells. Blood, 135(25), 2312–2325. https://doi.org/10.1182/blood.2019002102
- Muñoz-Escobar, J., Matta-Camacho, E., Kozlov, G., & Gehring, K. (2015). The MLLE domain of the ubiquitin ligase UBR5 binds to its catalytic domain to regulate substrate binding. Journal of Biological Chemistry, 290(37), 22841-22850. https://doi.org/10.1074/jbc.M115.672246
- “Real-Time PCR Reagents and Kits,” Thermo Fisher Scientific. [Online]. Available: https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/reagents-kits.html. [Accessed: Jul. 31, 2024].
- “CRISPR Technology Information,” Thermo Fisher Scientific. [Online]. Available: https://www.thermofisher.com/us/en/home/life-science/genome-editing/genome-editing-learning-center/crispr-cas9-technology-information.html. [Accessed: Jul. 31, 2024].
- “CRISPR Validated Protocols,” Thermo Fisher Scientific. [Online]. Available: https://www.thermofisher.com/us/en/home/life-science/genome-editing/genome-editing-learning-center/genome-editing-resource-library/crispr-validated-protocols.html. [Accessed: Jul. 31, 2024].
- S. A. Swenson et al., “UBR5 HECT domain mutations identified in mantle cell lymphoma control maturation of B cells,” Blood, vol. 136, no. 3, pp. 299-312, Jul. 2020. doi: 10.1182/blood.2019002102. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/32325489/. [Accessed: Jul. 31, 2024].
- “Know About Mantle Cell Lymphoma (MCL),” Calquence. [Online]. Available: https://www.calquence.com/why-calquence/know-about-condition-mcl.html. [Accessed: Jul. 31, 2024].
- “Additional Efficacy Data,” Tecartus. [Online]. Available: https://www.tecartushcp.com/car-t-cell-therapy/mantle-cell-lymphoma/additional-efficacy-data. [Accessed: Jul. 31, 2024].
- S. V. Forstater et al., “The Continuing Evolution of CAR T-Cell Therapy for B-Cell Malignancies,” The New England Journal of Medicine, vol. 387, no. 3, pp. 252-263, Jul. 2022. doi: 10.1056/NEJMra2202672. [Online]. Available: https://www.nejm.org/doi/full/10.1056/NEJMra2202672. [Accessed: Jul. 31, 2024].
Media Attributions
- further_enhanced_image
- Image 1
- Image 2

