Translation
Objective 6.4
6.4.1 Explain how mRNA is translated into protein.
A codon table is reproduced here. This is the molecular biologist’s Rosetta Stone, the mechanism by which an RNA sequence can be turned into a protein sequence.
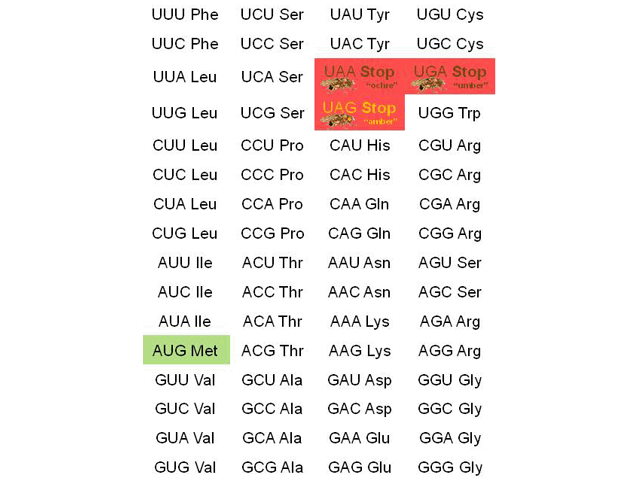
No one memorizes the codon table, and we don’t expect our students to do it, either. Rather, you need to be able to use the codon table and understand how it works.
Four codons deserve special mention. Remember that we need a starting place, a stopping place, and codes for the amino acids in between.
Protein synthesis always starts with the codon AUG, which codes for the amino acid methionine. For that reason, AUG is termed the start codon. If the final protein does not need methionine in this position, then it is cleaved away after the protein chain is made.
Protein synthesis ends with either UAA, UAG, or UGA. These are called the stop codons.
Oddly, each has its own name: UAA is called ochre; UGA is called umber; and UAG is called amber. It sounds like names from a bad spy movie. In reality, the names are fruit fly (Drosophila) eye colors. Fruit flies were an important part of genetic research for many years, and mutations to a UAA (where it shouldn’t be) produced flies with ochre eyes. Other eye colors were formed by mutations to the other stop codons.
RNA is translated into protein. Translation changes the language. In this case, translation changes the A, C, G, U language to the amino acid language. Four bases are translated into 20 amino acids.
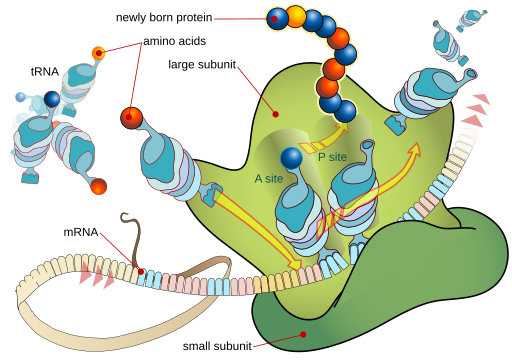
Protein synthesis occurs through the action of a macromolecular machine called a ribosome. Ribosomes can be found floating in the cytoplasm, not apparently attached to anything; in that case, they are called “free” ribosomes. Free ribosomes make proteins that the cell needs. The protein that is created is dumped directly into the cytoplasm.
Many ribosomes are associated with endoplasmic reticulum. In that case, the complex of ribosomes plus membranes is called rough endoplasmic reticulum (RER). The RER is continuous with the Golgi complex. The ribosomes, RER, and Golgi together make a system that is designed to make proteins for export, to be embedded in the cell membrane, or to recycle proteins that are malformed.
Also, proteins that need some sort of special processing (e.g. the addition of other molecules, such as sugars) go through the Golgi complex. Glycoproteins are usually, but not always, found on the cell surface.
Now we can see the special roles played by each type of RNA:
- Messenger RNA carries the coded message from the nucleus to the ribosomeThe ribosome is made up of ribosomal RNA and proteins. Together, rRNA and protein form ribosomes.
- The ribosome is made up of ribosomal RNA and proteins. Together, rRNA and protein form ribosomes.
- The factory is supplied with amino acids for protein synthesis by transfer RNA, the “trucks” that bring the correct amino acid for the next “word” in the genetic code. Each tRNA has a unique anticodon (a set of three ribonucleotides which will bind to the mRNA). This anticodon is paired with a specific amino acid which binds to an acceptor arm. For example, the anticodon 5’–GGG–3’ (complementary to the codon 5’–CCC–3’, as listed in the codon table) is found on a tRNA that is carrying the amino acid proline. These pairings are the basis for the genetic code.
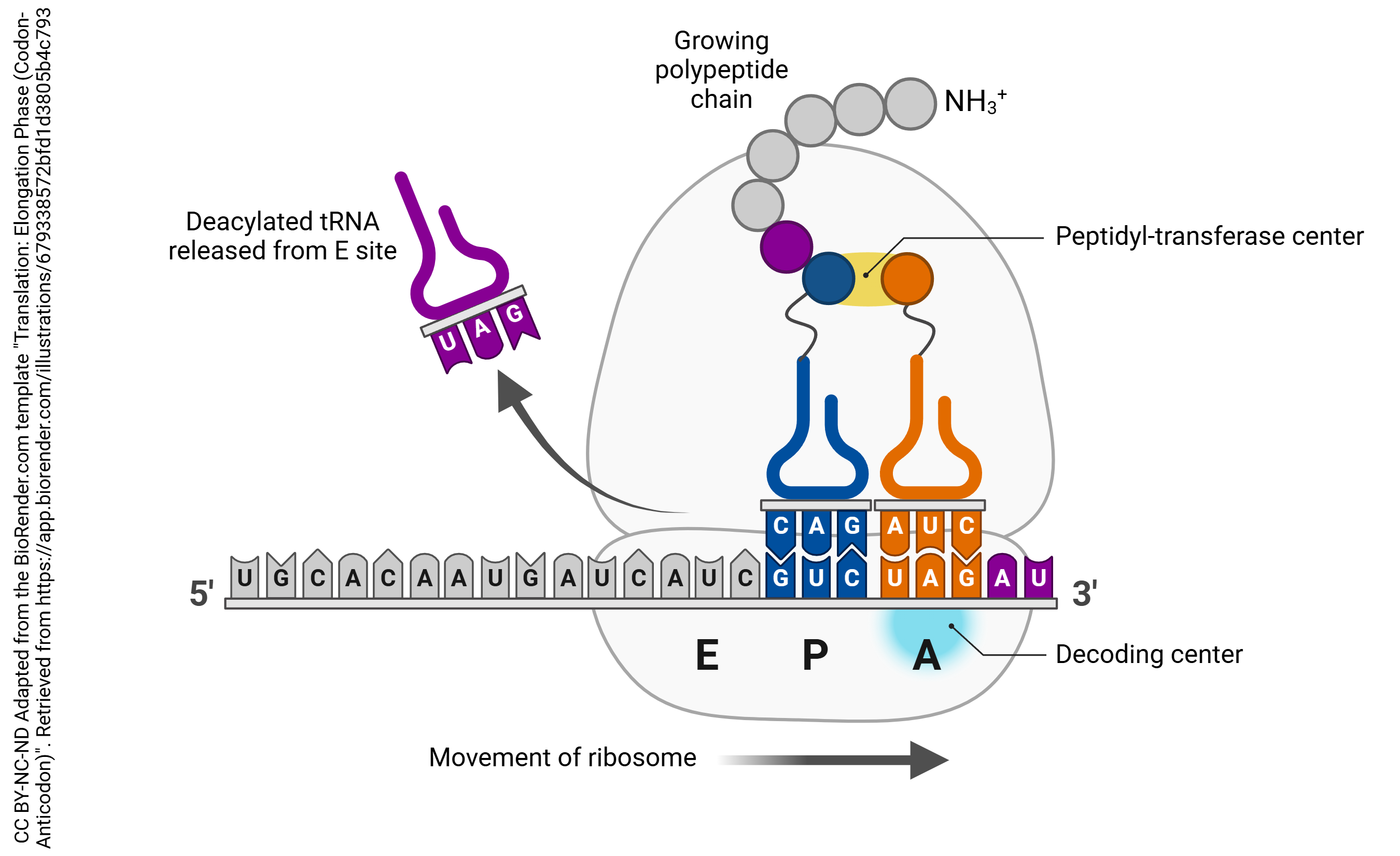
The large subunit of the ribosome has three special sites, termed “P”, “A”, and “E”. “P” and “A” stand for fairly complex molecular names, so don’t worry about them. Here is an easier way to think about it. The P site is where the growing polypeptide chain is made; the A site is where the new amino acids are added, and the E (exit) site binds to tRNA right before it leaves the ribosome. In other words, the A site is where single tRNAs carrying their amino acid cargo are docked. The P site is where the tRNA carrying the growing polypeptide chain is located. The details of protein synthesis by the ribosome follow.
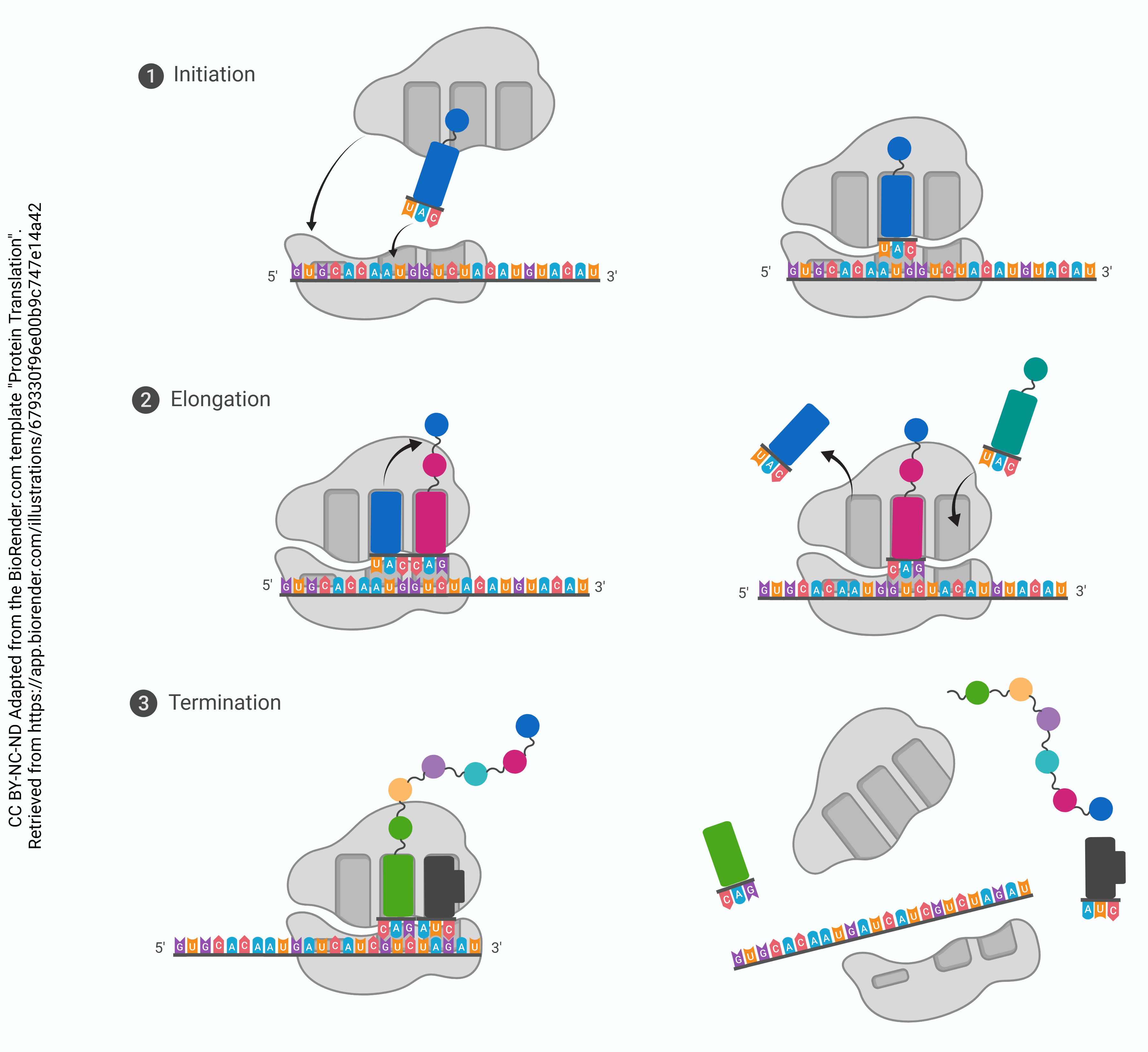
Stepwise Process for Translation:
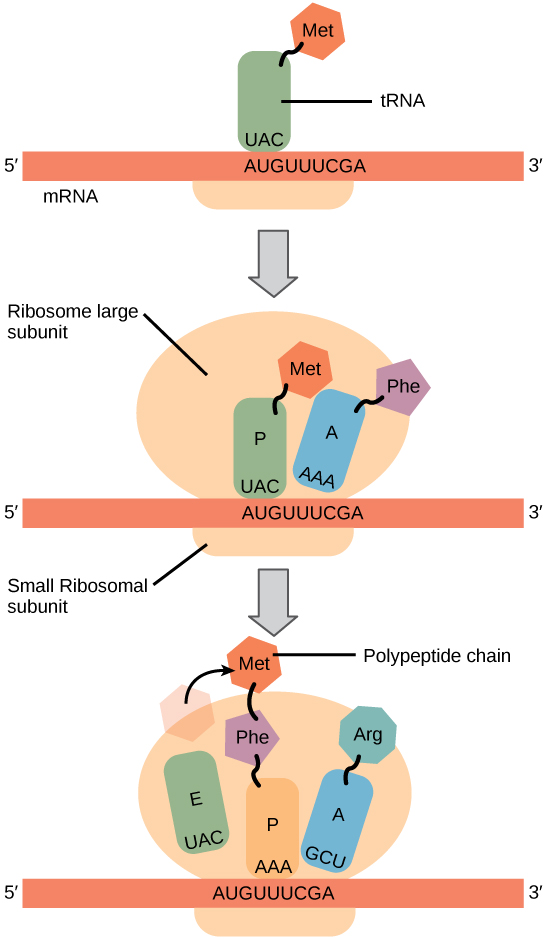
Step 1. mRNA attaches to the “P” position of the ribosome. 5’–AUG–3’ is always the start codon. Its complementary anticodon is 5’–CAU–3’. The tRNA with the anticodon CAU always carries a methionine.
When the AUG start signal is detected on the mRNA, Met-tRNA binds there.
Step 2. The binding of Met-tRNA triggers assembly of the small (40S) and large (60S) ribosomal subunits into a functional macromolecular machine at the site of tRNA binding. The large (60S) ribosomal subunit always assembles so that the Met-tRNA is enveloped in the P site. The A site is empty at this point.
Step 3. Now, the ribosome is assembled and ready to begin synthesis of the protein strand. The next codon is read (for example, AAU, which codes for asparagine or Asn). The Asn-tRNA nestles into the A site, next to the Met-tRNA which is still in the P site.
Step 4. In this step, the amino acids who find themselves snuggled together in the P and A sites decide they might as well form a peptide bond. When they do, the synthesis of protein has begun. The methionine is transferred to the Asn-tRNA and the tRNA which used to carry methionine is released to go get another one. This leaves the P site open.
Step 5. The Met-Asn-tRNA moves into the vacant P site as the ribosome ratchets three bases down (i.e., toward the 3’ end) the mRNA molecule. Now, the A site is empty. The next codon is (for example) 5’-UUA-3’. This codes for leucine, so the Leu-tRNA will be bound to the ribosome complex at that site.
Step 6. This process continues, and the polypeptide chain grows.
Step 7. The stop codon is reached. There is no tRNA for the stop codon; rather, the stop codon leaves the A site empty and the full-length protein is bound to the last tRNA at the P site. The ribosome disassembles, and the tRNA, protein, and ribosomal subunits all drift away from each other. The primary structure of the protein is now established.
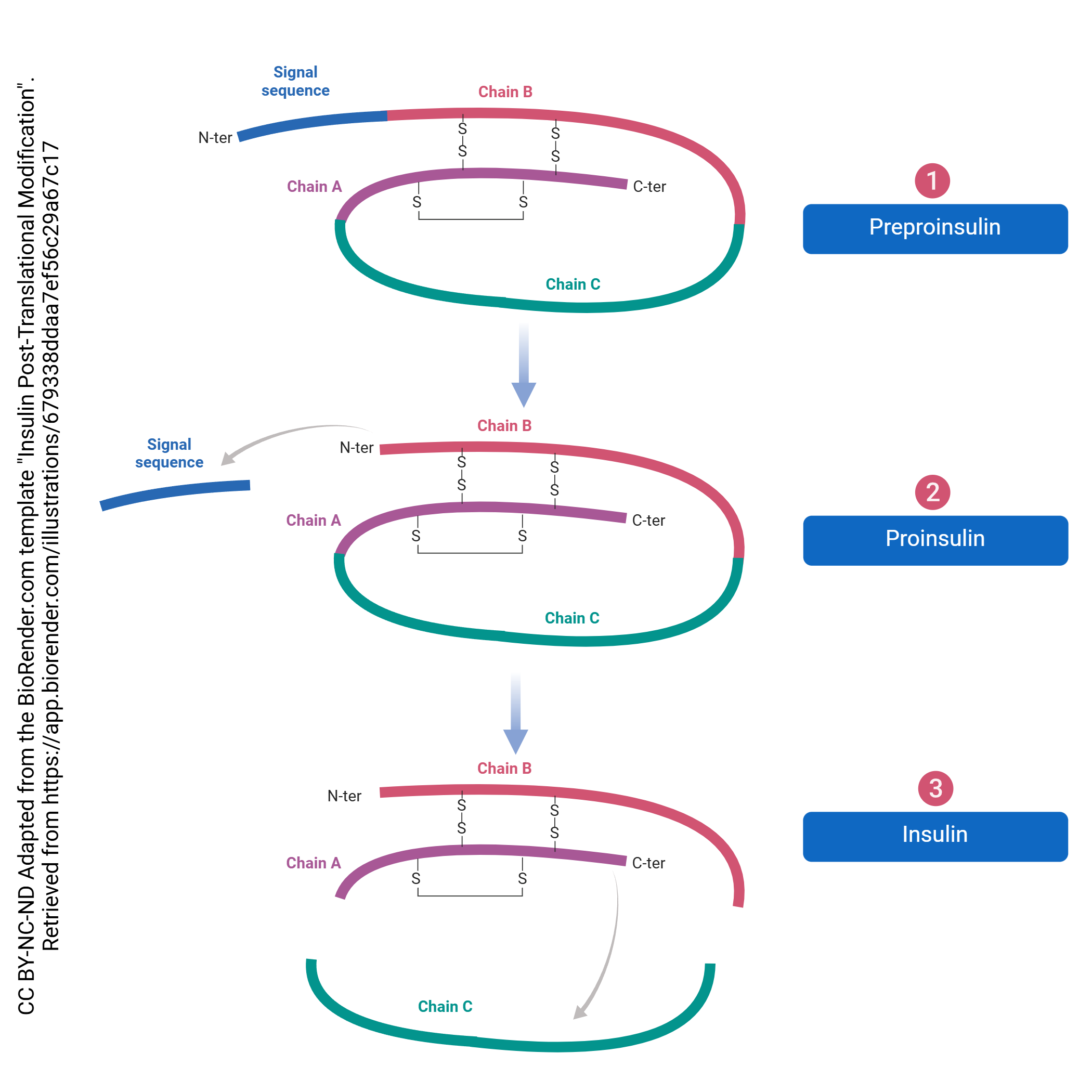
Just as there is processing of the pre-mRNA into the final mRNA, there is processing of the protein which must be done to create its final form. This processing begins in the RER and continues in the Golgi complex. For example, even though most proteins don’t start with methionine, all translation starts there. This is called post-translational processing because it happens after translation. The post-translational processing of insulin is shown here. It’s not important to know the details of this process for this specific signaling molecule, but rather get a sense of the general process that transforms the protein into its final, functional form.
Media Attributions
- U06-029 codon table 640x480px © Hutchins, Jim is licensed under a CC BY-SA (Attribution ShareAlike) license
- U06-014 Protein Translation © Elizabeth Rebarchik is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Ribosome_mRNA_translation_en.svg © LadyofHats is licensed under a Public Domain license
- U06-030 Translation_ Elongation Phase (Codon-Anticodon) © Samara Ona adapted by Elizabeth Rebarchik is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- OpenStax Translation © Betts, J. Gordon; Young, Kelly A.; Wise, James A.; Johnson, Eddie; Poe, Brandon; Kruse, Dean H. Korol, Oksana; Johnson, Jody E.; Womble, Mark & DeSaix, Peter is licensed under a CC BY (Attribution) license
- U06-031 Insulin Post-translational Modification © Qingyang Chen | BioRender is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license

