Synthesis Organelles
Objective 5.12
5.12.1 Name and classify the organelles involved in the synthesis of materials needed by the cell.
The synthesis organelles are thought of as macromolecular machines, collections of proteins and/or nucleic acids that work in concert to carry out complicated tasks inside the cell.
Two examples of these macromolecular machines we’ll discuss here are spliceosomes in the nucleus and ribosomes in the cytoplasm, although there are many, many others.
Spliceosomes
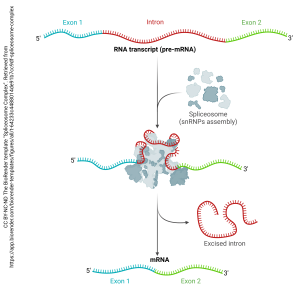 The spliceosome (“splice” + –soma, “body”) edits RNA copied from a DNA template and creates messenger RNA (mRNA) for export from the nucleus. The mRNA is then used as a template for protein synthesis.
The spliceosome (“splice” + –soma, “body”) edits RNA copied from a DNA template and creates messenger RNA (mRNA) for export from the nucleus. The mRNA is then used as a template for protein synthesis.
We’ll discover the details of this process in Unit 6.
Ribosomes
 Messenger RNA represents a “handoff” from one macromolecular machine (the spliceosome) to another (the ribosome). As the name suggests, the ribosome is a machine made up of ribonucleic acid (RNA) and protein. The specific type of RNA used is ribosomal RNA, or rRNA.
Messenger RNA represents a “handoff” from one macromolecular machine (the spliceosome) to another (the ribosome). As the name suggests, the ribosome is a machine made up of ribonucleic acid (RNA) and protein. The specific type of RNA used is ribosomal RNA, or rRNA.
Ribosomes read the mRNA template created by spliceosomes and translate it into a polymer of amino acids (a protein).
Ribosomes are therefore the machines that make proteins. We will study the details of this process in Unit 6.
Ribosomes can exist alone in the cytoplasm (free ribosomes) or associated with a membrane network (rough endoplasmic reticulum). Free ribosomes make proteins needed in the cytoplasm and some organelles. Rough endoplasmic reticulum-associated ribosomes are used to make proteins for export, proteins for display on the cell surface, or proteins that are destined for digestion organelles (Objective 13).
Endoplasmic Reticulum
The endoplasmic reticulum (endo–, “inside”; –plasm–, “mold”, “shape”, –ic; reticulum, “network”) is a complex network of membranes inside the cell. There are two types of endoplasmic reticulum seen in the electron microscope: without ribosomes (smooth endoplasmic reticulum, or SER) and with ribosomes (rough endoplasmic reticulum, or RER).
Smooth Endoplasmic Reticulum
 Smooth endoplasmic reticulum looks like one type of organelle under the electron microscope, but comprises at least three different functional types:
Smooth endoplasmic reticulum looks like one type of organelle under the electron microscope, but comprises at least three different functional types:
- SER for synthesis, discussed here;
- SER for storage (Objective 13)
- SER for digestion (Objective 13)
SER is used to synthesize steroids and other lipids. Mitochondria also aid in the synthesis of lipids, which is shown in the Canvas course but not discussed here in the textbook.
Rough Endoplasmic Reticulum
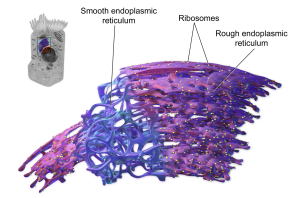
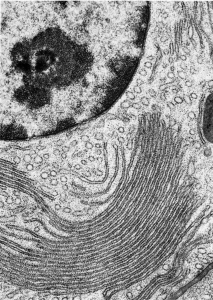
SER and RER are distinguished from one another by the absence (SER) or presence (RER) of ribosomes. The small, electron-dense ribosomes (which look like pepper grains in the EM) are what make RER “rough”.
The individual ribosomes associated with endoplasmic reticulum are apparent in this electron micrograph.
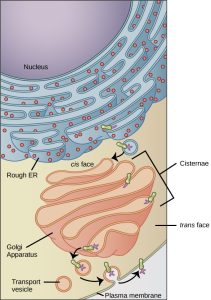
Golgi Apparatus
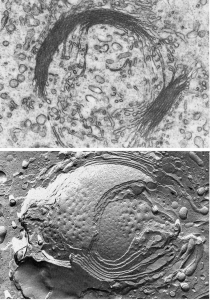
 Once a protein is made by the RER, it is modified by the Golgi complex (Golgi apparatus). Camillo Golgi, a 19th century Italian microscopist, is generally credited with its discovery although we didn’t figure out how it worked until after the electron microscope was invented in the second half of the 20th century.
Once a protein is made by the RER, it is modified by the Golgi complex (Golgi apparatus). Camillo Golgi, a 19th century Italian microscopist, is generally credited with its discovery although we didn’t figure out how it worked until after the electron microscope was invented in the second half of the 20th century.
Proteins which are processed in the Golgi apparatus have three possible fates:
- export from the cell via exocytosis;
- display on the surface of the cell;
- destruction inside a digestive organelle.
Proteins inside the Golgi apparatus are edited; refolded; and/or modified. Editing removes unneeded amino acids. Refolding then takes the edited protein, breaks old associations that hold the tertiary structure in place, and creates a new tertiary structure with new associations. Modifications can include hundreds of possible chemical reactions with wonderful names that we don’t need to know like farnesylation and geranylgeranylation. (These are fun to say at the dinner table, even if we don’t need to know them.)
There are two modifications that we do need to know and will use throughout the course: glycosylation and phosphorylation.
Glycosylation is the addition of a glycan moiety (sugar “tree”). Remember from Unit 4 that cells display their identity (“flags”) with glycoproteins on the cell surface.
Phosphorylation is a regulatory mechanism used to make proteins more, or less, active. The addition of a phosphate group is carried out by an enzyme called a kinase which will be a very important process that we’ll study extensively in the second semester (HTHS 2111), for example here, here, and here.
The Golgi apparatus takes in nascent (“newborn”) proteins at the cis or entry face (called cis because it’s closest to the RER and entry because it’s where incoming proteins arrive at the Golgi).
The Golgi spits out product at the trans or leaving face. There, it’s packaged into spherical membrane-bound vesicles (Latin: “little bladders”).
These vesicles can:
- fuse with the cell membrane and release their contents in the process called exocytosis we studied in Unit 4;
- fuse with the cell membrane which displays the glycan moieties on the outside of the cell;
- or remain as vesicles that become the digestion organelles we’ll study in Objective 13.
Misfolded or unneeded proteins are removed from the cell and recycled into amino acids by the last of these processes.
The Endomembrane System
It probably won’t surprise you that collections of macromolecular machines work together in a fashion akin to an industrial park. In the nucleus, the genetic material (DNA) is transcribed into RNA which is then edited into mRNA.
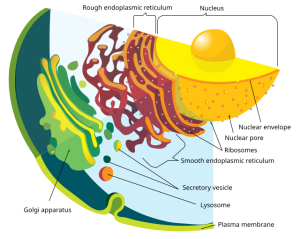
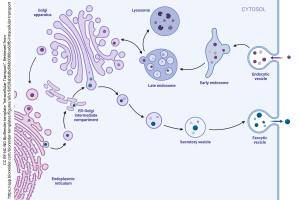
The mRNA is carried to the ribosome where it is made into proteins; if the ribosomes are associated with the endoplasmic reticulum, then vesicles carry the nascent protein to the Golgi apparatus. Different subregions of the Golgi carry out different sets of chemical reactions (as everyone knows, you don’t want to farnesylate where you geranylgeranylate). We shuttle proteins between these different machines within the Golgi complex using another set of vesicles called transfer vesicles.
Taken together, all these macromolecular machines, assembly lines, and packing/ shipping departments are called the endomembrane system.
Media Attributions
- U05-047 Spliceosome Complex © Hutchins, Jim is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- U05-048 Ribosome_shape © Vossman is licensed under a CC BY-SA (Attribution ShareAlike) license
- U05-049 smooth er © Kayser, Mike is licensed under a CC BY (Attribution) license
- U05-050 Figure_04_04_01 © Betts, J. Gordon; Young, Kelly A.; Wise, James A.; Johnson, Eddie; Poe, Brandon; Kruse, Dean H. Korol, Oksana; Johnson, Jody E.; Womble, Mark & DeSaix, Peter is licensed under a CC BY (Attribution) license
- U05-051 Blausen_0350_EndoplasmicReticulum © BruceBlaus adapted by Jordan West is licensed under a CC BY (Attribution) license
- U05-052 endoplasmic reticulum FawcettTheCellChapter5 p311 fig168 © Fawcett, Don is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- U05-053 FawcettTheCellChapter6 p378 fig197-198 © Fawcett, Don is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Endomembrane_system_diagram_en.svg © LadyofHats is licensed under a Public Domain license
- U05-055 Intracellular Transport © Hutchins, Jim is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license

