Replication | Duplication of DNA
Objective 6.5
6.5.1 Describe the process of DNA replication.
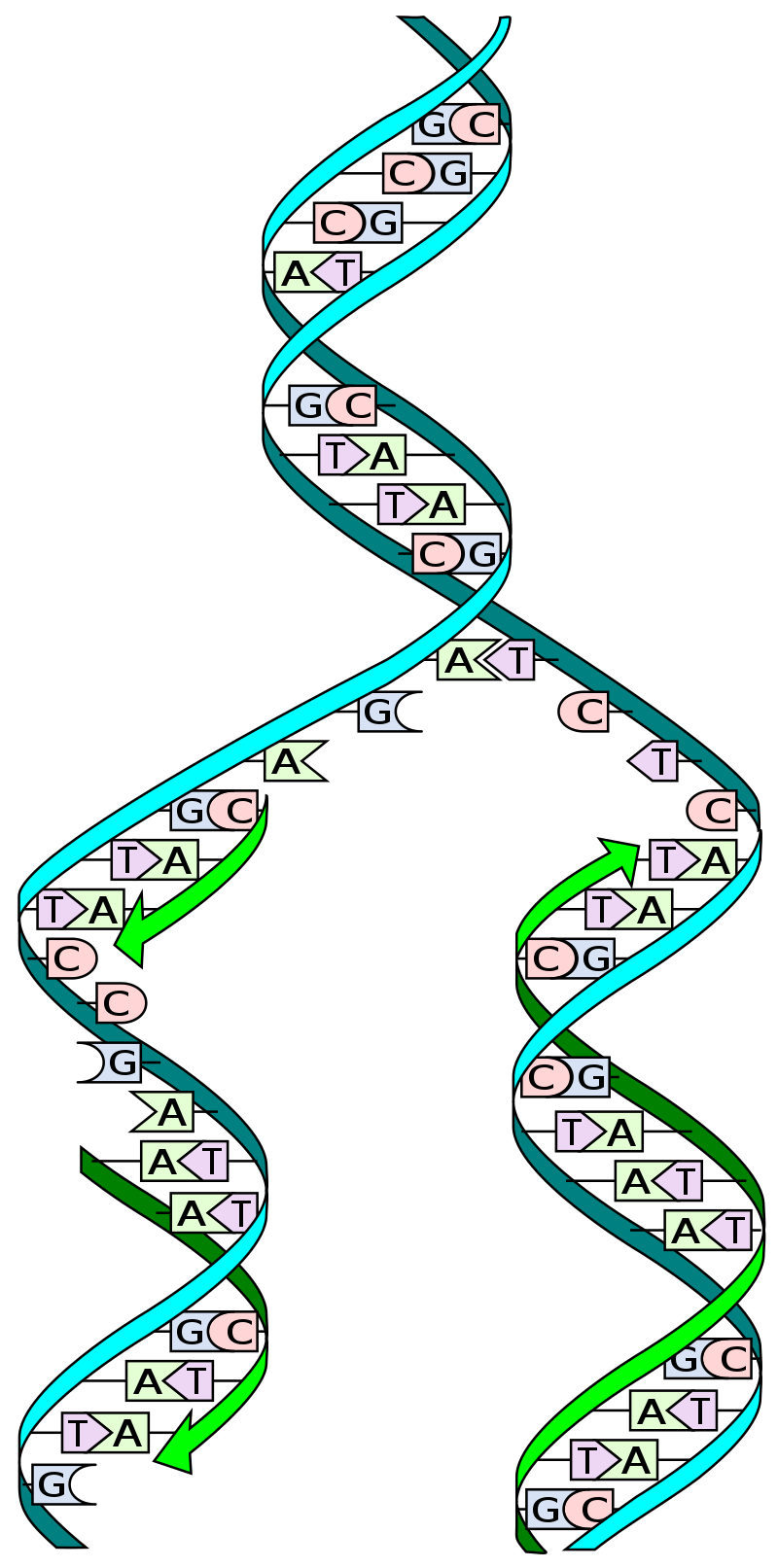
Dividing cells pass through a repeating series of steps called the cell cycle, described in detail in Objective 6. DNA replication, the copying of all the cell’s DNA molecules, occurs once during each cycle of cell division. The ability of DNA to replicate with high fidelity is one of its key features. In this way, the genetic material is passed from cell to cell and from generation to generation.
DNA is replicated during the synthesis (S) phase of the cell cycle.
Each chromatid is one continuous DNA molecule wrapped around histones to form chromatin. A normal human cell (not involved in sexual reproduction) has 46 DNA molecules. During S phase, DNA is copied in a process called replication. Each strand of DNA is used as a template to make a new strand of DNA, so that the amount of DNA in the cell is doubled. The number of DNA molecules increases to 92. These 92 molecules are packed into 46 chromosomes (each with two chromatids, so 2 x 46 = 92). When the DNA is divided in half, each daughter cell gets 46 DNA molecules.
As described earlier, Watson and Crick immediately perceived that the structure of DNA they proposed led directly to a mechanism of doubling DNA. This was one of the most beautiful and powerful aspects of their proposed model. Scientists prefer hypotheses which have strong explanatory power and this was one of the purest examples of a hypothesis which opened up an entirely new field, what the philosopher and historian of science Thomas Kuhn called a paradigm shift. The field their discovery created is called molecular biology.
DNA is only synthesized in the 5’ to 3’ direction, and can only be built from a single-stranded template. The double helix is opened into two strands, one running 3’ to 5’ and the other antiparallel strand running 5’ to 3’. The opening is called the replication fork.
This creates a problem for the cell, because there are two antiparallel strands to be copied by different mechanisms. The strand that opens into the fork in the 3’ to 5’ direction can be copied directly by building the antiparallel 5’ to 3’ strand quickly. Because this process happens fast, that strand of the new double helix is called the leading strand.
For the strand running 5’ to 3’, special techniques, which take longer, are needed to copy it. This copied strand is called the lagging strand.
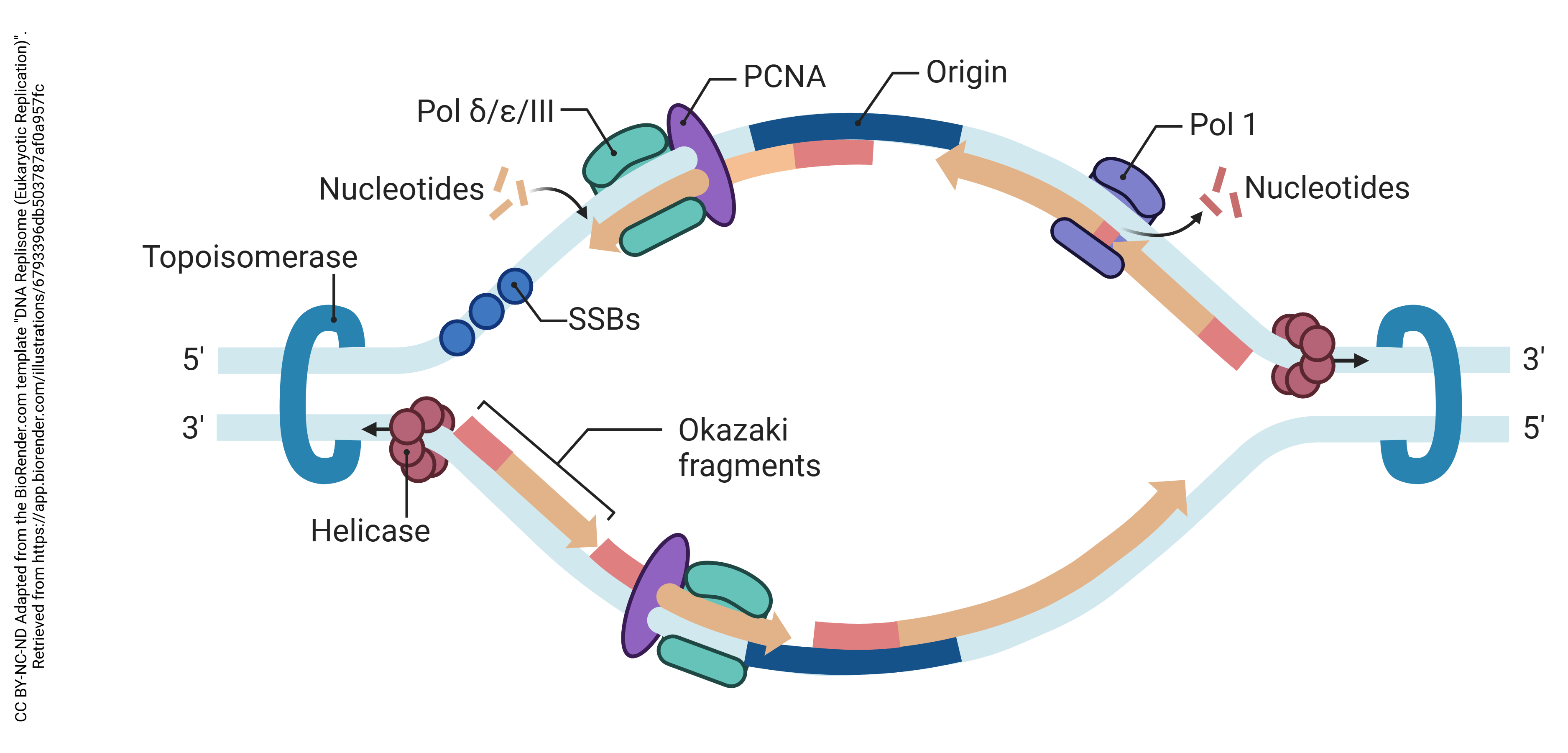
The machinery needed for DNA replication is shown in the diagram.
An enzyme called topoisomerase acts as a swivel to relieve the stress created from the unwinding of individual DNA strands. If you can imagine tying a rope to a fixed point, then pulling on the strands of the rope to separate them, you will quickly understand the problem that topoisomerase solves. If you can’t visualize it, this video (also in the Canvas course) might help.
Helicase separates the two strands of the DNA double helix at the split in the replication fork.
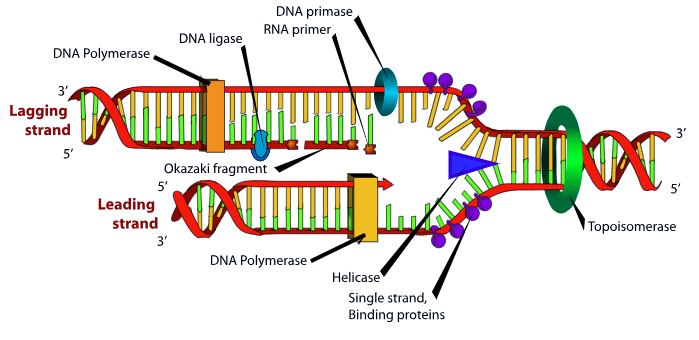
The leading strand is built by a DNA polymerase which slides in the 3’ to 5’ direction and builds new DNA in the 5’ to 3’ direction (left to right in this diagram). DNA polymerase requires a short double helical segment to bind to DNA. If a scientist needs to start DNA polymerase, they will add a short single-stranded DNA called a primer which is complementary and antiparallel to the target DNA sequence to create this short double-stranded region for DNA polymerase to start with. The newly formed strand provides this primer function.
The lagging strand has to be built in smaller pieces. We have to wait for enough of the replication fork to open up in order for DNA polymerase to get access to it and build right to left in the diagram above. The lagging strand opening continues until the single-stranded DNA is a few hundred bases in length. Then, primase, an RNA polymerase, lays down an RNA primer. The DNA polymerase, moving right to left in the diagram, builds a double-stranded region a few hundred bases long in the usual 5′ to 3′ direction until it is stops when it reaches the previously-built double-stranded sequence. These double-stranded DNA-DNA hybrid regions, several hundred bases long, are called Okazaki fragments. The RNA primers are removed and the Okazaki fragments are stitched together later, but this process is not shown in the diagram.
All this to-ing and fro-ing takes a considerably longer time than just making DNA, so it’s easy to see why the lagging strand takes longer to make.
Media Attributions
- U06-032 DNA_replication_split.svg © Madprime is licensed under a CC0 (Creative Commons Zero) license
- U06-034 DNA Replisome (Eukaryotic Replication) © BioRender is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- DNA replication © LadyofHats Mariana Ruiz adapted by Elizabeth Rebarchik is licensed under a Public Domain license

