College of Science
98 Explicating the Substrate-engaged Signals for Type 3 secretion of FlgJ in Salmonella enterica
Julianna Matteis; Kelly Hughes; and Fabienne Chevance
Faculty Mentor: Kelly Hughes (School of Biological Sciences, University of Utah)
ABSTRACT
The bacterial flagellar motor is representative of one of the most vital micro- mechanical machines in biology. Flagellar rotation allows cells, in this case Salmonella enterica bacteria, to navigate and move towards nutrients or away from toxic substances and facilitates bacterial pathogenesis. The flagellum structure is commonly divided into three parts: a basal body, hook, and filament. Biosynthesis of the bacterial flagellum in Salmonella enterica depends on the secretion and assembly of numerous proteins for its construction, including the subject protein of this study, FlgJ. Many of the proteins required for the structure and assembly of the flagellum are secreted from the cytoplasm by a flagellar-specific type III secretion (T3S) apparatus assembled at the cytoplasmic base of the structure. A related T3S system in Salmonella is used to construct a hypodermic-like structure related to the flagellar basal structure, called the injectosome. Effector proteins are secreted through the injectosome into host cells by Salmonella and other infectious bacteria to facilitate pathogenesis.
A component of the flagellar basal body is the rod structure. The rod is divided into proximal and distal rod half-structures. The rod acts as a drive shaft and assembles in the periplasmic space from the secretion apparatus within the cytoplasmic membrane through the cell wall to the outer membrane. The proximal rod polymerizes to the cell wall after which the FlgJ cap is added to the tip of the completed proximal rod. FlgJ serves two roles. FlgJ has muramidase activity and effectively chews a hole in the cell wall. FlgJ is also a scaffold that allows distal rod subunits to assemble beneath it until the rod reaches the outer membrane. Through a series of mutagenesis techniques, the characteristics of the rod cap protein, FlgJ, could be explored to help understand its role in the type III secretion system.
The research conducted in this study utilized a fusion of β-lactamase (Bla) to the cap FlgJ to both select and quantify secretion. Bla is normally secreted into the periplasm through the cell’s general secretion system called the Sec secretion pathway. Once in the periplasm, Bla folds into an active conformation where it confers resistance to beta- lactams such as ampicillin. If the Sec secretion signal is removed from the Bla protein, it will not get secreted into the periplasm, and cells are ampicillin-sensitive (ApS). However, Bla lacking its sec secretion signal can be fused to secreted flagellar proteins, and if a Fla-Bla chimera is secreted into the periplasm via the flagellar T3S system, it will fold into an active conformation and confer ampicillin resistance (ApR) to the cell.
My research utilized a genomic editing method known as λ-red recombination to engineer FlgJ-Bla fusions with parts of FlgJ deleted to determine which residues of FlgJ were needed for FlgJ-Bla secretion. A genetic method, phage transduction, was used to move genetic markers from one strain to another to make genetic combinations of alleles.
I was able to make a series of 10 amino acid, in-frame, deletions of a FlgJ-Bla fusion, which could then be tested for their ability to be secreted in different genomic backgrounds. Phenotype characteristics such as motility and minimal inhibitory concentrations (MICs) to ampicillin resistance allowed me to determine if the FlgJ-Bla constructs were functional for FlgJ activity and how efficiently they were secreted into the periplasm. One significant finding resulting from this research was the discovery that the fusion of β-lactamase (Bla) to full-length FlgJ (FlgJ-Bla strain) did not interfere with FlgJ function. The FlgJ-Bla fusion allowed normal flagellum assembly as the bacteria expressing a FlgJ-Bla fusion exhibited wild-type motility. The characterization of deletion alleles of the FlgJ-Bla fusion allowed for the identification of specific amino acid (AA) signals that target FlgJ for secretion. This will allow future researchers to understand how the type III secretion system recognizes the amino acid signals that target substrates for
INTRODUCTION
Each year, millions of individuals are infected with Salmonella worldwide, consequently leaving many of those infected suffering from physiological distress. Salmonella enterica, a gram-negative, prokaryotic bacterium can elicit high levels of virulence, despite it being only microns in size. Its rod-shaped body and helical extensions, called flagella, allow the bacteria to propel itself effectively in liquid environments, which also facilitates pathogenesis. The corkscrew motility of counterclockwise spinning flagella allows the bacteria to propel themselves forward in different liquid environments, whereas clockwise rotation allows the bacterium to change direction if the cell senses it is going in an unfavorable direction. Approximately 70 genes comprise the flagellar regulon, creating a nanomachine with complex morphology (Chevance et al., 2017). The structural components of the flagella can be discerned into three portions: the basal body, a membrane-embedded protein that spans from the cytoplasm to the outer membrane; a ~55 nm flexible hook that protrudes from the outer membrane and attaches to the hook-filament junction; and the filament, which extends off the hook, whose rotation propels the Salmonella enterica cell. A flagellar-specific type III secretion apparatus assembles at the cytoplasmic base of the structure. Numerous proteins are required to complete flagellum assembly that are secreted from the cytoplasm within the cell. Something particularly unique about this type III secretion system (T3S) is its ability to differentiate specific proteins as early and late secretion- substrates. Within the structure of this nanomachine, the hook-basal body and rod are composed of early secretion-substrates, while the filament is a by late secretion-substrate. When the hook reaches its terminal length the flagellar T3S apparatus undergoes a secretion-specificity switch from early to late secretion-substrate selectivity.
The genes of the flagellar regulon are expressed in a transcriptional hierarchy of three classes of promoters: Class I, Class II, and Class III. Their expression is coupled to flagellum assembly. The products of the class I promoter genes (flhCD) facilitate RNA polymerase recognition of class II promoter genes. Class II promoters direct transcription of genes needed for the construction of the hook-basal body (HBB). The formation of the HBB structure is instigated by the formation of the MS-ring, composed of FliF protein subunits, within the cytoplasmic membrane. Following the formation of the MS-ring, the C-ring, which is in the cytoplasm, is formed from proteins FliG, FliM, and FliN, which create the rotor of the flagellar motor. Following rod assembly, described earlier, the structures of the P-ring and L-ring are formed respectively around the distal rod to make a bushing in the outer membrane (Daofeng et al., 2022). During HBB assembly, the protein FliK acts as a molecular ruler be taking temporal measurements of the length of the growing structure as it is secreted out of the cell. This protein controls the length of the hook and is secreted before the filament substrates (Minamino et al., 2009). Once the hook reaches the expected length of ~55 nm, FliK catalyzes the substrate-specificity of the secretion apparatus and instigates late substrate secretion mode. FlgM, which inhibits the class 3 promoter specific transcription factor, 28, prior to HBB completion is a late secretion substrate. As FlgM is secreted, σ28 is free to transcribe the late substrate- encoding class 3 genes.
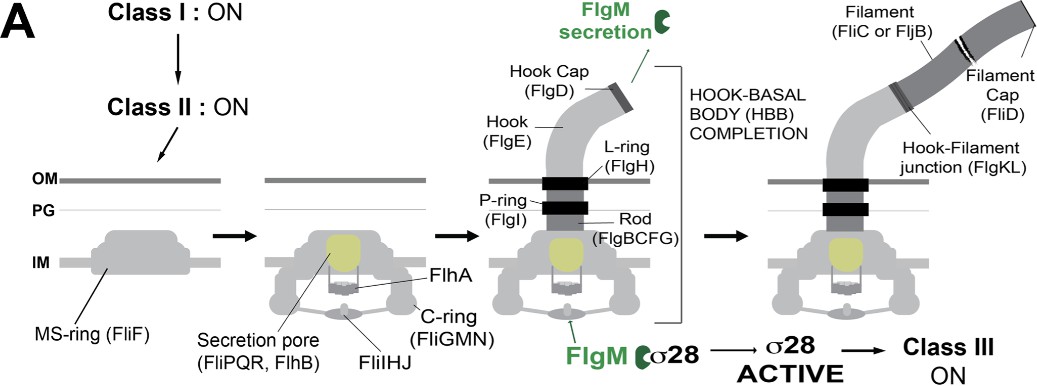
Figure 1. Illustrating the Flagellar Type III Secretion System (Doafeng et al., 2022).
To understand how the FlgJ protein is secreted from the cytoplasm, one must understand how it is recognized by the flagellar type III secretion (T3S) system for targeted secretion. The hook-basal body (HBB) structure of the flagellum acts as the flagellar motor onto which the long external filament polymerizes. The HBB contains a driveshaft, called the rod. The rod assembles within the periplasmic space, which is the region between the inner and outer membranes of Gram-negative bacteria. A proximal rod assembles first from the inner membrane to the cell wall. The protein FlgJ can be classified as a rod-cap and assembles itself on the tip of the proximal rod. Its muramidase activity digests a hole in the cell wall, allowing the distal rod to assemble. Two crucial signals in FlgJ can be found in both the N-terminal and C-terminal domains. The N- terminus’ role is to facilitate polymerization to allow the distal rod to be built, while the C- terminus creates a hole in the cell wall to advance assembly (Cohen et al., 2014). Before the distal portion of the structure is complete, FlgJ must be dislodged, which is believed to be done by the protein FlgD, which is the hook cap protein. Based on previous research, it was noted that FlgD was continuously secreted into the periplasm during PL-ring formation, which supports that FlgD is the determining factor for dislodging FlgJ at the time the L-ring is assembled. The synthesis of the L-ring thus removed the hook-cap FlgJ (Cohen et al., 2014). The distal rod is composed of FlgG protein subunits, and these polymerize beneath the FlgJ cap until the structure reaches the outer membrane.
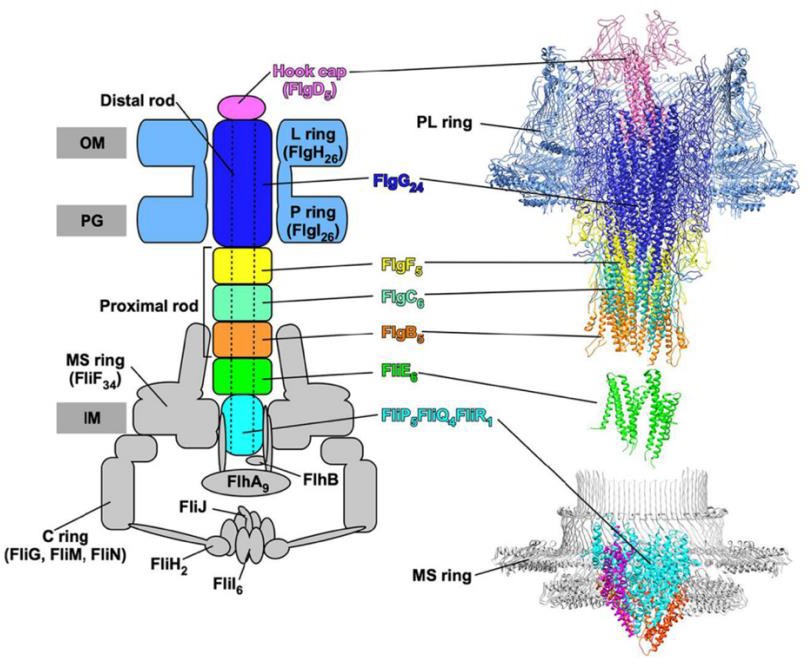
Figure 2. Salmonella enterica Basal Body Structure (FlgJ located in “Distal rod”) (Hendriksen et al. 2021)
Fusionofβ-lactamasetoFlgJ:
To study the secretion dynamics of flagellar proteins, β-lactamase was used as a reporter enzyme throughout the experiment. This approach leverages the fusion of β-lactamase (Bla) to target flagellar proteins, enabling quantifiable detection of any secretion within the cell. The gene encoding β-lactamase was fused to the gene encoding the flagellar protein of interest, in this case, FlgJ, which allows the production of a fusion protein that retains both flagellar functionality and β-lactamase activity. Bla must be secreted into the periplasm to fold into an active conformation where it will then cleave β-lactams, such as ampicillin, and confer antibiotic resistance (Chevance et al., 2023). Without its Sec- secretion signal, Bla is not secreted, but if fused to a flagellar secretion signal then Bla is secreted through the flagellar T3S system and results in ampicillin resistance (ApR).
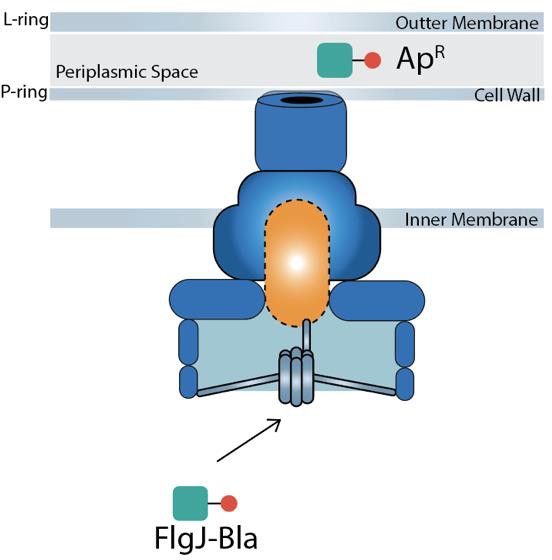
Figure 3. The secretion of FlgJ-Bla into the periplasm and folding into an active conformation, conferring ampicillin resistance.
A characteristic of FlgJ that becomes of interest in this proposal is the finding that the fusion of β-lactamase (Bla) to FlgJ (FlgJ-Bla strain) does not interfere with FlgJ function to prevent flagellum assembly as the bacteria-expressing a FlgJ-Bla fusion shows wild-type motility, which provided an experimental phenotype for FlgJ function. When testing the FlgJ-Bla strain on a motility plate, it was surprising to see that it had a motile function unlike its FlgB-Bla and FlgC-Bla counterparts, which are found in the rod component of the flagellar system but exhibited non-motile phenotypes.
MATERIALS AND METHODS
Bacterial strains and Media
Strains used in this study were derived from Salmonella enterica serovar Typhimurium wild-type strain LT2. Cells were cultured in lysis broth (LB: Bacto-tryptone 10g/L, Bacto- yeast extract 5g/L, NaCl 5 g/L). Antibiotics were added to LB at the following final concentrations: 100 µg/ml sodium ampicillin (Ap), 12.5 µg/mL chloramphenicol (Cm), 15 µg/mL tetracycline-HCl (Tc), 50µg/ml kanamycin sulfate (Km), 100 µg/mL carbenicillin, as needed. Apex agar (12g/L) was used for the preparation of a solid medium.
Insertion of the pSIM5 Plasmid
The pSIM5 plasmid encodes functions needed for Lambda-Red dependent genome editing. To introduce the pSIM5 plasmid into recipient cells, a donor strain containing the pSIM5 plasmid. The donor strain was grown in LB broth at 30°C (pSIM5 is temperature- sensitive for replication) in the shaking incubator until cell growth reached late log phase. P22, a bacteriophage, was used to infect the Salmonella enterica cells for the transduction process. Transduction is the process where infection phage such as P22 will package non-P22 DNA and introduce it into newly infected cells. P22-H5, a lytic phage, was used after cells were purified following transduction to be sure they can be re- infected. This screens against P22 phage resistant transductants following purification. The recipient strain, TH24148 was grown in an LB culture at 37°C until an OD of ≈ 0.4-0.6. The recipient strain was infected with the P22 in a 1:10 dilution and incubated at 30°C for 60 min before plating to allow for expression of pSIM5-encoded chloramphenicol resistance. 100 µL of the incubated culture, alongside the diluted phage stock, was plated on a green plate and incubated overnight. Using a 0.1 mL pipette, a line of H5 phage lysate was applied down the middle of a green plate and allowed time for the liquid to soak in. Single, light-green colored colonies were picked from the green plates were streaked across H5 to check for P22-sensitivity as part of the post-transduction purification process. Once colonies were purified, they were streaked on LB-Cm plates and kept at 4°C.
FlgJ tetRA Cassette
The tetRA cassette is a tetracycline-resistance element from transposon Tn10. The TetA protein is an efflux pump in the inner membrane that pumps tetracycline from the cytoplasm. The TetR protein is a transcriptional repressor that inhibits its own expression and tetA expression in the absence of tetracycline. The tetRA cassette is a useful genetic tool in that it can be selected for and against. Insertion of tetRA into the Salmonella chromosome confers resistance to tetracycline (TcR), but the presence of the TetA pump in the membrane makes the cells sensitive to lipophilic toxic compounds such as fusaric acid. Thus, cels expressing TetA will die on fusaric acid-containing medium, which we call tetracycline-sensitive (TcS) medium because it is a selection for loss of the tetRA element. The strain TH24148 contains the flgJ-bla fusion. This strain was used as the wild-type precursor from Salmonella enterica where tetRA marker was inserted replacing 10 amino acids in the flgJ coding sequence, creating 10 equal-sized segments, that of which were deleted by oligonucleotide-directed genome editing replacement of the tetRA cassettes via -Red recombination. PCR products were purified via ethanol precipitation. PCR program: tetRA-Phusion program
98° C – 2 minutes for 1 cycle
98° C – 30 seconds, 56° C – 30 seconds, 72° C – 1 minute 10 seconds for 30 cycles
72° C – for 2 minutes for 1 cycle
λ-Red Recombination
Red recombination is an efficient way of targeting distinct chromosomal locations for genome editing. Using only a small portion of the 5’ end of the targeted DNA sequence in Salmonella enterica, specific recombination proteins facilitate the recombination at those sequences (Karlinsey, 2007). Exonucleases are used to degrade the 5’ ends of the linearized DNA, creating single-stranded portions ready to be targeted for recombination. A binding protein attaches itself to the single-stranded region and invades any homologous regions within the plasmid. Gam, which in an inhibitor of RecBCD, prevents degradation of the single-stranded portions while the exonucleases generate the 3’ ends which will be fully recombined (Karlinsey, 2007). Plasmid pSIM-5 (CmR) or pSIM-6 (ApR), encodes for the λ-Red recombinase and was used experimentally to facilitate λ-Red targeted mutagenesis for FlgJ. The pSIM plasmids contains a temperature-sensitive origin of replication and thus was grown at the permissive temperature of 30°C, in the presence of Ap or Cm. The λ-Red functions are induced via a temperature-inducible promoter. The selection of tetracycline-sensitive (TcS) recombinants was done on zinc- fusaric acid selection media, which were used for further experimentation.
Main procedure used for λ -red recombination during this research is as outlined:
- Cells were grown in 1 mL culture at 30°C overnight
- Subculture 1:100 culture diluting 250 µL of the overnight culture in 25 mL of LB media. Grow the cells at 30° C until it reaches an OD of approximately 0.2
- Heat shock cells in the 25 mL flask at 42° C for 15 minutes to induce the l-red proteins
- Ice the cells for 5-10 mins
- Transfer the cells to a 50 mL falcon tube
- Spin down cells at max speed for 10 minutes using a centrifuge
- Discard supernatant, ensuring that the pellet is undisturbed
- Add 25 mL of ice MQ water and resuspend cells via vortexing
- (Repeat steps f-h)
- Following the discarding of the supernatant the third time, vortex the pellet to ensure that all cells are resuspended in the remaining water in the Falcon tube. Ensure that cells are always on ice during transition periods.
- Electroporate the tetRA-PCR (100ng up to 300ng) into 50 µL of washed cells. Using the correct electroporation programs, electroporate cells alongside a no- DNA cell control.
- Immediately after electroporation, add 800 µL of LB media into the cuvette and resuspend electroporated cells by pipetting up and down and transfer the cells to a culture tube.
- Incubate all culture tubes at 37° C for 45 minutes
- Plate 300 µL of each culture on LB, TcS, and incubate overnight at 37° C
- The following day, streak 12 independent, TcR colonies for isolation on LB plates at a temperature of 42° C
- Test the 12 colonies previously picked on TcS plates, and LB plates and incubate at 37° C
- Circle on LB plates which colonies are TcS on the TcS plates. (Can test other antibiotics using this method.
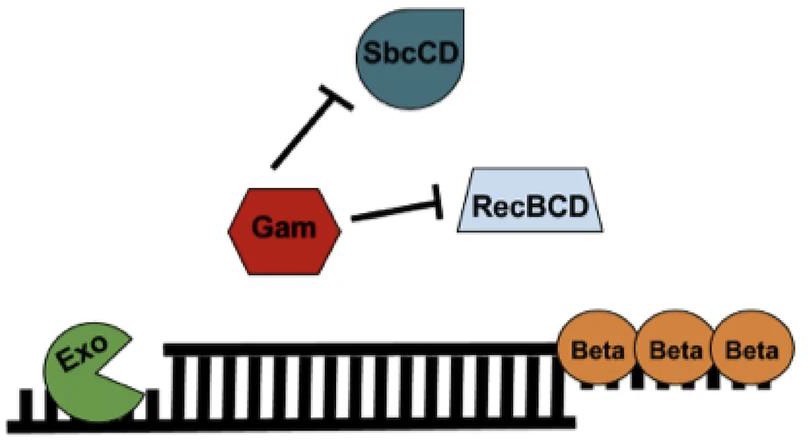
Figure 5. Components of the Lambda-Red Recombineering system (Kenkel, 2016).
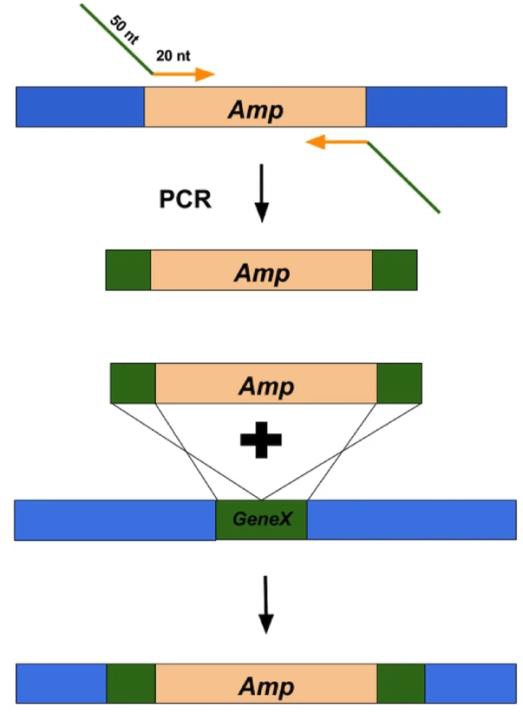
Figure 6. Overview of using Lambda-Red Recombineering system to replace a gene of interest with an antibiotic resistance cassette (Kenkel, 2016).
Sequencing of FlgJ-Bla deletions:
Following λ-Red mutagenesis, mutants were grown in LB overnight cultures. Cultures were diluted using sterile water and boiled at 96° C for 10 minutes to be used as a template in a PCR reaction. The PRC product was subject to agarose gel electrophoresis, confirming that the small DNA fragments of the correct size were obtained. Purification of the PCR products was completed by a Qiagen purification kit before being sent off for Sanger Sequencing analysis. NCBI Nucleotide Blast, a computer program, was used to analyze the chromatogram and confirm that the clean deletions of FlgJ were in the correct base pair position without undesired mutations.
Construction of Strain in ΔflgBC Background
Construction of FlgJ strains were selectively tightened in the ΔflgBC background (which excludes the rod and hook component of the flagellar structure). This was experimentally achieved using transduction, a technique used in the Hughes’ lab (see instructions for integration of pSIM5 plasmid).
FlgJ-Bla Motility Assays
Experimentation was done to test the bacterial motility (Mot+) of strain TH24148 (FlgJ- Bla) grown from freezer stocks, streaked on fresh LB plates to form single colonies which would then be poked into soft-agar motility plates. The degree of the motility was compared to that observed for the wild-type Mot+ strain, LT2. Meanwhile, I tested the minimum inhibitory concentration (MIC) to ampicillin for the strain expressing the FlgJ-Bla fusion constructs. The parental FlgJ-Bla expressing strain formed colonies on protease peptone bile-salts (PPBS) Ap15R plates and was motile.
Screening of FlgJ mutants in ΔflgBC Background:
All FlgJ strains used during experimentation were fused to β-lactamase, to detect the level of secretion. If FlgJ-Bla was secreted into the periplasm of the cell, it would fold into an active conformation and confer ampicillin resistance. The level of ampicillin resistance for each of the FlgJ deletions in a ΔflgBC background was determined using a minimum inhibitory concentration assay (MIC). Further analysis was done via a Western Blot to determine the effects of the deletion constructs on protein expression or stability to recognize if there were any important residues in the regions of FlgJ including amino acids 20-50. Colonies from both ΔflgBC mutants and flgB+C+ were evaluated for ability to assemble into a functional structure, which is determined as able to secrete the FlgM protein and transcribe Class III genes. Tetrazolium-Lactose plates (Tz-Lac) and MacConkey Lactose (Mac-Lac) plates were utilized to determine class 3 promoter expression levels in strains expressing the lac operon from a class 3 flagellar promoter. Assays on the Mac-Lac plates will present a color gradient from a white, to light pink, to pink, to red with increasing lac operon expression. Colonies that are pink to red in color will demonstrate that lactose is being fermented, while colonies that display little color indicate that lactose is not being fermented in the cell and class 3 promoter express is off indicating that the HBB was not assembled. On the other hand, Tz-Lac plates display an inverted color pattern compared to Mac-Lac. White colonies would suggest that the gene expression of the strain is high, while a red color suggests that the overall class 3 promoter expression is low.
RESULTS
ΔflgBC MIC assays
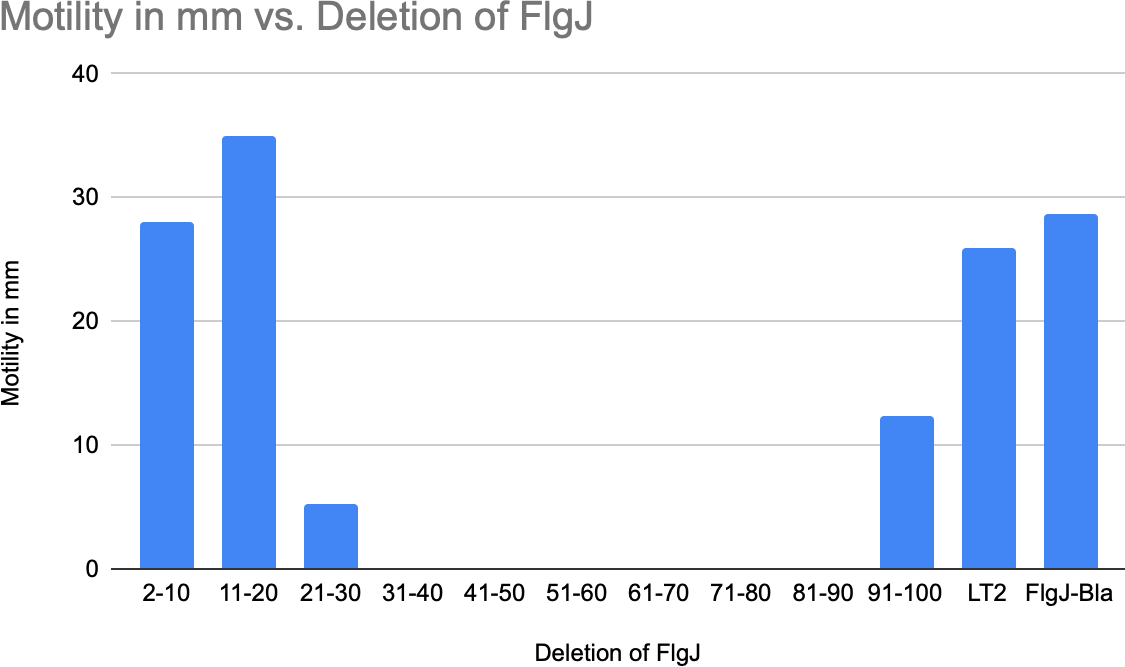
Effect of FlgJ deletions on FlgJ-Bla secretion in the periplasm (in a rod mutant background):
|
FlgJ deletion |
Ampicillin Resistance via MIC (ApR under 2-fold dilutions) |
|
TH29213 (Δ amino acids 2-10 of FlgJ) |
6 |
|
TH29151 (Δ amino acids 11-20 of FlgJ) |
6 |
|
TH29152 (Δ amino acids 21-30 of FlgJ) |
0 |
|
TH29153 (Δ amino acids 31-40 of FlgJ) |
12.5 |
|
TH29154 (Δ amino acids 41-50 of FlgJ) |
3 |
|
TH29155 (Δ amino acids 51-60 of FlgJ) |
6 |
|
TH29156 (Δ amino acids 61-70 of FlgJ) |
12.5 |
|
TH29157 (Δ amino acids 71-80 of FlgJ) |
12.5 |
|
TH29158 (Δ amino acids 81-90 of FlgJ) |
12.5 |
|
TH29286 (Δ amino acids 91-100 of FlgJ) |
12.5 |
|
TH29159 (full-length FlgJ) |
12.5 |
|
LT2 (strain lacking the FlgJ-Bla fusion) |
0 |
An MIC (Minimum Inhibitory Concentration) assay was done to determine the lowest concentration of ampicillin was required to inhibit the growth of FlgJ-Bla expressing strains in the ΔflgBC background. This assay provides important information about the effectiveness of specific deletions on FlgJ-Bla secretion.
Tz-Lac and Mac-Lac Agar Assays
|
ΔflgBC background |
Mac-Lac Assay |
Tz–Lac |
|
TH29213 (Δ 2-10) |
Pink/Red |
Light Pink |
|
TH29151 (Δ11-20) |
Pink/Red |
Light Pink |
|
TH29152 (Δ21-30) |
Pink/Red |
Light Pink |
|
TH29153 (Δ31-40) |
Light Pink |
Dark Red |
|
TH29154 (Δ41-50) |
Light Pink |
Dark Red |
|
TH29155 (Δ51-60) |
Light Pink |
Dark Red |
|
TH29156 (Δ61-70) |
Light Pink |
Dark Red |
|
TH29157 (Δ71-80) |
Light Pink |
Dark Red |
|
TH29158 (Δ81-90) |
Light Pink |
Dark Red |
|
TH29286 (Δ91-100) |
White/Pink |
Pink |
|
TH29159 |
Dark Pink |
Light Pink |
|
LT2 |
White |
Dark Red |
FlgJ-BlaMotilityAssaysResults(Photographmissing2-10,91-100)
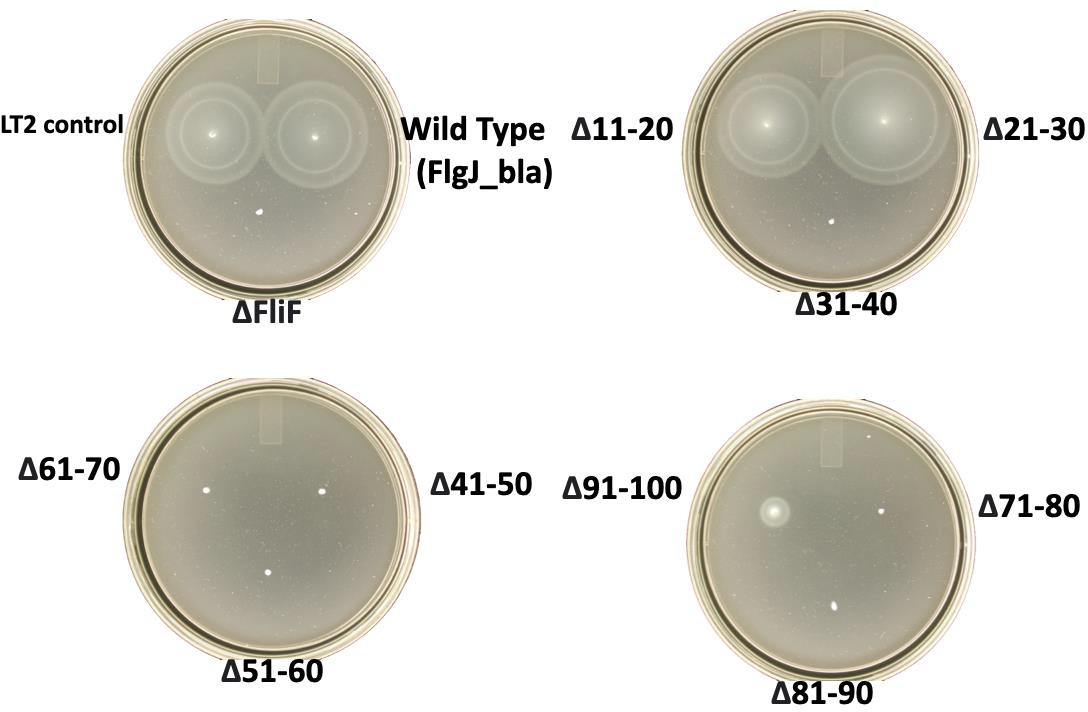
DISCUSSION
This research project was designed to explore regions within the first 100 residues of the FlgJ protein of Salmonella enterica that would affect its ability to function in HBB formation or its ability to be recognized as a substrate for the flagellar T3S system. FlgJ has an integral role within the flagellar system, acting as a scaffold capping protein for flagellar distal rod formation. Research has shown that the function of FlgJ is to facilitate the polymerization of the distal rod, creating a hole in the peptidoglycan layer (cell wall) during flagellum assembly. This process allows for the rod component of the flagellar system to elongate to the outer membrane of the cell, advancing the assembly of the basal body (Hirano et al., 2001).
By constructing a series of deletions in flgJ coding for regions within the first 100 amino acids in a FlgJ-Bla fusion construct, critical regions were determined in FlgJ for it to be properly secreted and assembled in the flagellar system. Even though specific amino acids were pinpointed in this research, our results demonstrate how an earlier signal in amino acids 21-30 may affect the secretion dynamics of FlgJ, compared to another early secretion substrate, FlgB, that had been previously characterized. The Hughes’ lab found that phenylalanine residue F45 of FlgB was essential for its secretion. FlgJ also contains phenylalanine at amino acid 44, which I initially thought would be important for secretion. Deletion of residues 41 – 50 prevented FlgJ-Bla secretion, which would be consistent with F44 being an important residue. This would have to be verified by specifically targeting the F44 residue to determine effects of single amino acid substitutions at residue 44 on FlgJ-Bla secretion.
The deletion of 21-30 amino acid region in FlgJ (TH29152) resulted in a complete loss of ampicillin resistance, suggesting that this region in the sequence plays a key role in the secretion of FlgJ, provided that the FlgJ-Bla protein lacking residues 21-30 is stable. Reduced secretion in other regions such as 31-40, and 41-50 align more with the findings with a the similar FlgB protein, but implicate that there is a more complex, nuanced role of how FlgJ interacts within the T3SS. Again, this would depend on whether these FlgJ-Bla constructs are stable.
The proteins that comprise the flagellum must successfully be secreted and assembled to result in a motile phenotype and to facilitate pathogenesis. FlgJ, unlike other Bla fusion proteins in the structure, remained motile with a fusion to Bla. The fusion of FlgJ-Bla allowed us to assay for secretion, as it only requires a small amount of protein for the rapid response of secretion to take place, but the motility assays exhibited the functional consequences of FlgJ secretion. For example, the segments of deletions in the central and C-terminal regions of FlgJ exhibited wild-type motility, despite the varying levels of ampicillin resistance. This observation notes that FlgJ’s ability to facilitate flagellar assembly and confer motility may not be directly tied to is efficient secretion as previously presumed. However, the phenotypic differences observed from the motility of TH29153 and TH29154 suggest that there may be downstream effects on flagellar function.
The screening of the FlgJ mutants on Mac-Lac and Tz-Lac agar plates allowed for further insight into the secretion dynamics of FlgJ. The phenotypic analysis on the Mac- Lac agar revealed that strains with mutations in the C-terminal region (e.g. TH29286, TH29159) displayed a shift in colony color from pink to white, suggesting a defect in lactose fermentation, which may have a dependable showing of flagellar disfunction. The defect in lactose fermentation may highlight proper flagellum assembly, hindering the ability of these mutants to navigate the medium and form colonies. Interestingly, colonies that were white or light pink on the Mac-Lac assays showcased a different level of secretion activity on the Tz-Lac assays, exemplifying the connection secretion and function of the flagellar system. Further experimentation will be done to evaluate the level of expression within the specific segments of deletions.
This research provides valuable insights into the complex regulation of the proteins that comprise the flagellar system in Salmonella enterica, highlighting regions of FlgJ that are important for its recognition and transport through the T3SS. The finding that certain deletions do not completely reduce the motility or secretion implies that there may be a level of flexibility in the system, which will require the study of other proteins in the T3S system to fully understand secretion-targeting signals. Further experimentation of FlgJ will help to narrow in on specific amino acid residues that define FlgJ’s secretion signal. Additionally, future research could expand on the use of β-lactamase fusion within the T3S system to study the dynamics of other flagellar proteins within the structure, or even in broader applications regarding bacterial virulence.
In summary, this work contributes to the growing understanding of how bacteria regulate flagellar assembly and secretion using small tools, such as FlgJ, embedded within their systems. To have a better understanding on the world around us, one should look more closely and notice the unseeable mechanisms that have a big impact. This research was not only about the story of FlgJ, but rather how such small things, like proteins and cells, can hold so much character, offering new avenues for the study of bacterial properties and pathogenesis.
APPENDIX
FlgJ deletions in wild-type background:
|
Strain Number |
Chromosome Genotype |
Operation |
|
TH29059 (Δ 2-10) |
ΔflgJ9253::tetRA(deletion of a.a. 2-10) flgJ8650::bla |
λ-Red |
|
TH28440 (Δ 11-20) |
ΔflgJ9253::tetRA(deletion of a.a.11-20) flgJ8650::bla |
λ-Red |
|
TH28664 (Δ 21-30) |
ΔflgJ9274::tetRA(deletion of a.a. 21-30) flgJ8650::bla |
λ-Red |
|
TH28657 (Δ 31-40) |
ΔflgJ9273::tetRA(deletion of a.a. 31-40) flgJ8650::bla |
λ-Red |
|
TH28572 (Δ 41-50) |
ΔflgJ9256::tetRA(deletion of a.a. 41-50) flgJ8650::bla |
λ-Red |
|
TH28573 (Δ 51-60) |
ΔflgJ9275::tetRA(deletion of a.a. 51-60) flgJ8650::bla |
λ-Red |
|
TH28574 (Δ 61-70) |
ΔflgJ9258::tetRA(deletion of a.a. 61-70) flgJ8650::bla |
λ-Red |
|
TH28575 (Δ 71-80) |
ΔflgJ9259::tetRA(deletion of a.a. 71-80) flgJ8650::bla |
λ-Red |
|
TH28577 (Δ 81-90) |
ΔflgJ9261::tetRA(deletion of a.a. 81-90) flgJ8650::bla |
λ-Red |
|
TH28576 (Δ 91-100) |
ΔflgJ9260::tetRA(deletion of a.a. 91-100) flgJ8650::bla |
λ-Red |
|
TH 29213 (Δ 2-10) |
ΔflgBC6557 ΔflgJ9253::tetRA(deletion of a.a. 2-10) flgJ8650::bla |
T |
|
TH 29151 (Δ 11-20) |
ΔflgBC6557 ΔflgJ9253::tetRA(deletion of a.a.11-20) flgJ8650::bla |
T |
|
TH 29152 (Δ 21-30) |
ΔflgBC6557 ΔflgJ9274::tetRA(deletion of a.a. 21-30) flgJ8650::bla |
T |
|
TH29153 (Δ 31-40) |
ΔflgBC6557 ΔflgJ9273::tetRA(deletion of a.a. 31-40) flgJ8650::bla |
T |
|
TH29154 (Δ 41-50) |
ΔflgBC6557 ΔflgJ9256::tetRA(deletion of a.a. 41-50) flgJ8650::bla |
T |
|
TH29155 (Δ 51-60) |
ΔflgBC6557 ΔflgJ9275::tetRA(deletion of a.a. 51-60) flgJ8650::bla |
T |
|
TH29156 (Δ 61-70) |
ΔflgBC6557 ΔflgJ9258::tetRA(deletion of a.a. 61-70) flgJ8650::bla |
T |
|
TH29157 (Δ 71-80) |
ΔflgBC6557 ΔflgJ9259::tetRA(deletion of a.a. 71-80) flgJ8650::bla |
T |
|
TH29158 (Δ 81-90) |
ΔflgBC6557 ΔflgJ9261::tetRA(deletion of a.a. 81-90) flgJ8650::bla |
T |
|
TH 29286 (Δ 91-100) |
ΔflgBC6557 ΔflgJ9260::tetRA(deletion of a.a. 91-100) flgJ8650::bla |
T |
ACKNOWLEDGEMENTS
I would like to thank Kelly T. Hughes and Fabienne F.V. Chevance for guiding me through this research, as well as the other members involved in Hughes’ lab that helped create this story of FlgJ.
Bibliography
Chevance, F., & Hughes, K. (2023). Β-lactamase (BLA) reporter-based system to study flagellar type 3 secretion in Salmonella. BIO-PROTOCOL, 13(12). https://doi.org/10.21769/bioprotoc.4696
Cohen, E. J., & Hughes, K. T. (2014). Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. Journal of Bacteriology, 196(13), 2387–2395. https://doi.org/10.1128/jb.01580-14
Hendriksen, J. J., Lee, H. J., Bradshaw, A. J., Namba, K., Chevance, F. F., Minamino, T., & Hughes, K. T. (2021). Genetic analysis of the Salmonella FliE protein that forms the base of the flagellar axial structure. mBio, 12(5). https://doi.org/10.1128/mbio.02392-21
Hirano, T., Minamino, T., & Macnab, R. M. (2001). The role in Flagellar Rod Assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. Journal of Molecular Biology, 312(2), 359–369. https://doi.org/10.1006/jmbi.2001.4963
Karlinsey, J. E. (2007).‐Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods in Enzymology, 199–209. https://doi.org/10.1016/s0076- 6879(06)21016-4
Kenkel, B. (2016). LAMBDA Red: A homologous recombination-based technique for genetic engineering. Addgene blog. https://blog.addgene.org/lambda-red-a- homologous-recombination-based-technique-for-genetic-engineering
Minamino, T., Moriya, N., Hirano, T., Hughes, K. T., & Namba, K. (2009). Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Molecular Microbiology, 74(1), 239–251. https://doi.org/10.1111/j.1365-2958.2009.06871.x
Qu, D., Jiang, M., Duffin, C., Hughes, K. T., & Chevance, F. F. V. (2022). Targeting early proximal-rod component substrate FlgB to FlhB for flagellar-type III secretion in Salmonella. PLOS Genetics. https://doi.org/10.1371/journal.pgen.1010313

