Spencer Fox Eccles School of Medicine
59 Intracellular Amino Acid Accumulation in Cells Grown in Amino Acid Deficient Growth Media and the Role of Ornithine and Polyamines in the Growth of ASS1 Deficient Cancer Cells
Austin Bender; Kyle Dunlap; and Gregory Ducker
Faculty Mentor: Gregory Ducker (Biochemistry, University of Utah)
In the process of tumorigenesis, metabolic pathways are often rewired to shunt molecules away from non-essential processes and towards proliferation1. A frequent adaptation in the development of melanoma is the down regulation of argininosuccinate synthetase 1 (ASS1), the rate limiting enzyme in the conversion of citrulline and aspartate to the amino acid arginine (Arg) (Figure 1A and 1B)2. Growth of ASS1 deficient cells is dependent upon media Arg (Figure 1C)3,4. Consequently, depletion of Arg is being evaluated as a potential therapy for the treatment of ASS1 deficient cancers4-6. However, clinical efforts to exploit this vulnerability have been predominantly unsuccessful, potentially due to the re-expression of ASS1 in cells exposed to an Arg depleted environment over time7-9. We sought to discover new approaches that would synergize with Arg starvation to kill cells. To do this, we engineered an ASS1 knockout in the A375m melanoma cell line using CRISPR-Cas9.
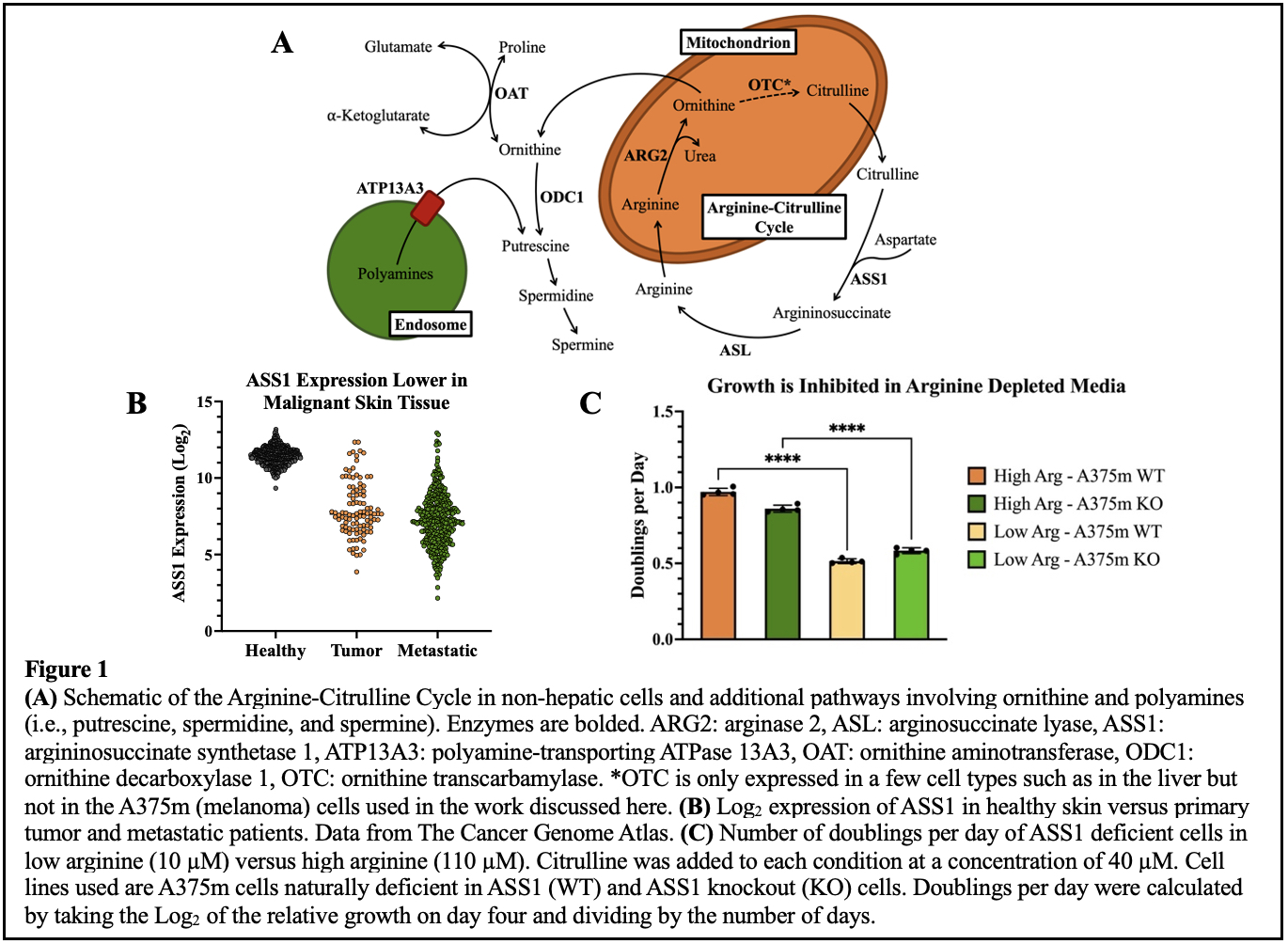
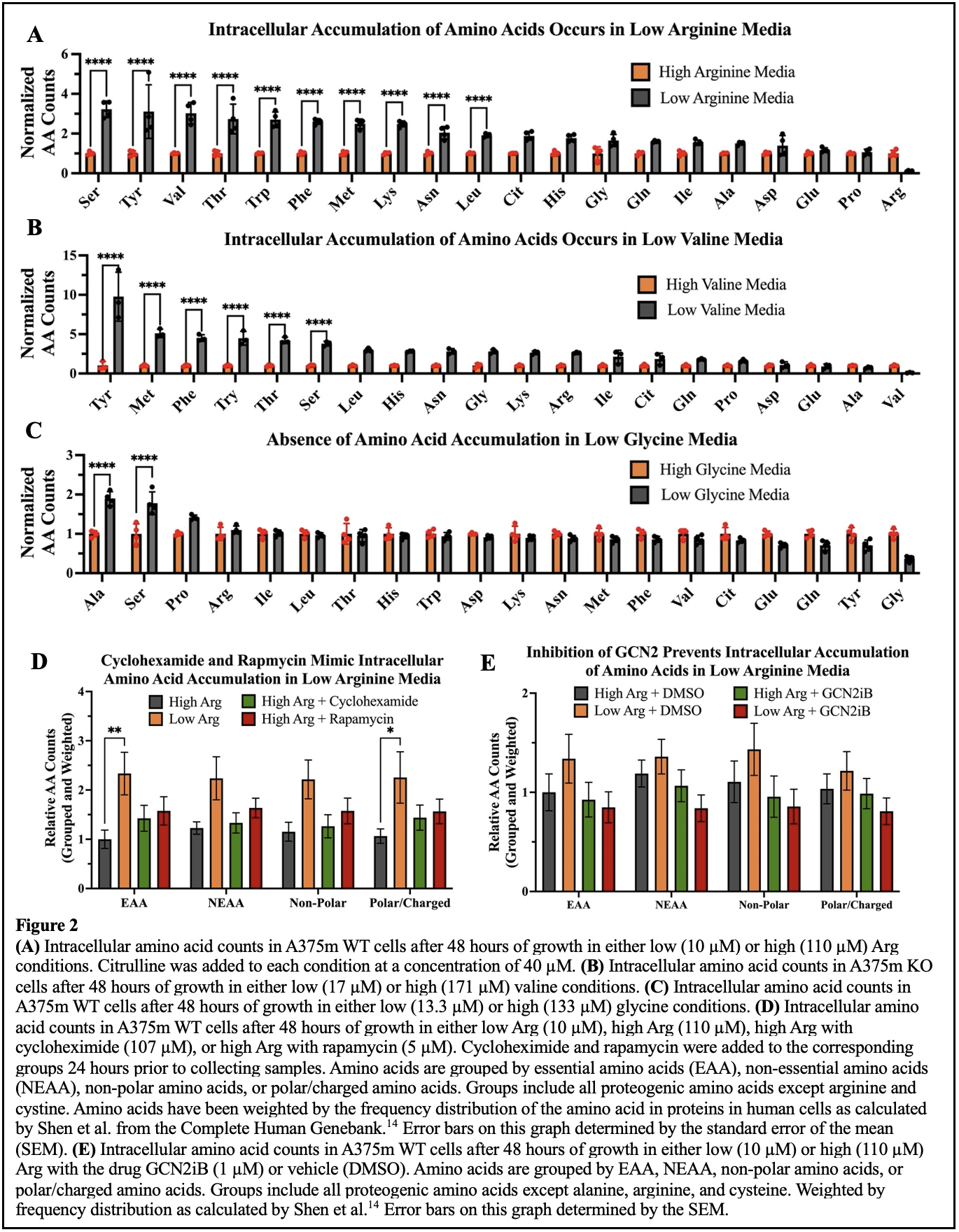
However, in characterizing the behaviors of our cell lines cultured in low arginine media, we uncovered a counterintuitive increase in intracellular metabolites. In low Arg (10 µM) conditions, A375m cells accumulated more intracellular amino acids when compared to cells grown with high Arg (110 µM) (Figure 2A). To understand what cells were responding to that induced this change, we then altered the restricted amino acid. We observed amino acid accumulation in media depleted of the essential amino acid valine, but not of the non-essential amino acid glycine (Figure 2B and 2C). This led us to conclude that an accumulation phenotype was in response to an essential amino acid limitation. Next, we asked if this phenotype was the result of a translational dependent process. To test for translational control over amino acid accumulation, we conducted an experiment that treated ASS1 deficient cells with either rapamycin, an inhibitor of the nutrient sensing complex mTOR which controls translation of nutrient utilization and sensing pathways, or cycloheximide, an inhibitor of protein synthesis10,11. The results were then compared to cells grown in Arg depleted conditions. Both the inhibition of mTOR and translation resulted in a similar amino acid accumulation as observed in the cells deprived of Arg which suggests a translation-related mechanism (Figure 2D).
Based on known mechanisms linking amino acid starvation and mTOR signaling, we proposed that amino acid accumulation was induced by the general control nonderepressible 2 (GCN2) pathway. GCN2 is activated when the cell is starved of proteogenic amino acids leading to the suppression of mTOR activity and the phosphorylation of eukaryotic initiation factor 2 alpha (eIF2a) which inhibits the translation of most mRNAs aside from those which might help the cell adapt to the amino acid deficiency12,13. We then asked whether an inhibitor of GCN2 would block this metabolic phenotype. Indeed, GCN2 inhibition caused a reversal of the phenotype suggesting that it plays a role in the intracellular accumulation of amino acids that is observed when Arg is depleted from the cellular environment (Figure 2E).
From these experiments, we learned more details about our model system and how cells in general can respond to an amino acid deficiency. Because the amino acid accumulation phenotype was only observed in response to restriction of an essential amino acid, we conclude that the cell likely makes the deficient amino acids when it can do so, such that it is no longer in deficiency, and the GCN2 pathway is not activated in response. In the case of ASS1 downregulation, the cell losses its ability to produce endogenous Arg causing what is typically a non-essential amino acid to become essential to the cell. When extracellular Arg becomes low, ASS1 deficient cells can respond by activating the GCN2 pathway to stave off cell death, and therefore this pathway might serve as a target for future treatments of ASS1 deficient cancers.
Having gained a better understanding of the pathway leading to the amino acid accumulation phenotype observed, we then performed a functional genomics screen using CRISPR to identify synthetic lethalities occurring with loss of ASS1 and Arg starvation. To do this we grew cells in low Arg media, knocked out a pool of metabolism-related genes using CRISPR, and then allowed the cells to grow for roughly 15 generations. Total guide RNA abundances were quantified to establish which gene knockouts had a growth phenotype. This was done with both A375m ASS1 KO cells and A375m cells which have been genetically engineered to have ASS1 overexpressed (ASS1 OE). This comparison allowed us to find genes that when knocked out are particularly detrimental to the cell in combination with loss of ASS1 in low Arg conditions. A top gene hit from the screen encodes for the mostly endosome localized, ATPase 13A3 (ATP13A3) polyamine transporter (Figure 1A)15-17. As a downstream product of Arg catabolism, polyamines have essential roles in cellular functions including RNA splicing and nucleic acid stabilization18. Therefore, we hypothesized that ASS1 deficient cells grow poorly in Arg depleted conditions due to a lack of polyamines and significantly reduced growth will result with the added loss of ATP13A3.
To test our hypothesis, we grew cells with lacking ASS1 in increasing Arg concentrations with putrescine (Put), a polyamine, added to standard RPMI media. We found improved growth with Put supplementation with 20 – 110 µM Arg. Additionally, Put caused no growth improvement in cells with ATP13A3 knocked out, which supports its function as a polyamine transporter as the cells with ATP13A3 knocked out were likely unable to access the Put from the media. Interestingly, the improved growth seen with ornithine (Orn), a precursor to polyamines and other metabolites (e.g., proline and glutamate)19, was greater than that seen with Put (Figure 3A). However, Orn alone could not rescue the growth of cells plated in media completely void of Arg which suggests that Orn is not acting as a replacement for Arg within the cell but is beneficial via some other pathway (Figure 3B). This suggests that polyamine deficiency is not the primary factor that inhibits the growth of ASS1 deficient cells in low Arg environments. Furthermore, Orn’s failure to rescue growth in the complete absence of Arg confirms that the cell is missing an enzyme needed to make Arg from this substrate. However, the added growth in the presence of limited Arg suggests that a non-Arg pathway is involved in the cell’s utilization of the supplemental Orn to increase growth. Therefore, the metabolic versatility of ornithine presents as a point worthy of future inquiry.
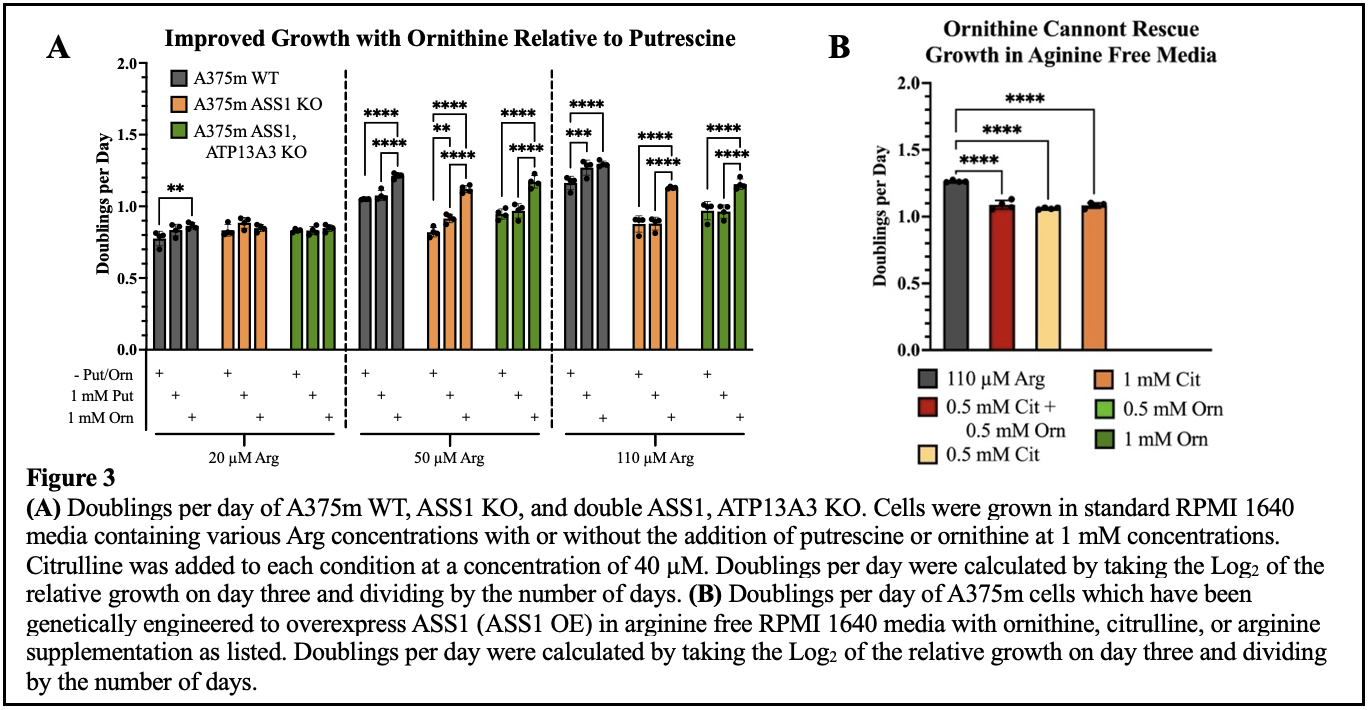
If ASS1 is down regulated in the process of tumorigenesis, what was once a non-essential amino acid to the cell, Arg, becomes essential and uptake of exogenous sources is crucial for survival. When the environment is depleted of Arg, our findings show a GCN2 mediated pathway is likely responsible for slowing translation which restricts cellular growth and likely provides the cell enough time to adapt to the new conditions. Subsequently, the GCN2 pathway might serve as an additional target in coordination with the Arg depletion therapies currently being evaluated. We hypothesize that inhibition of GCN2 will render the short-term adaptability of the cell less effective and thus delay long term adaptation through re-expression of ASS1 by preventing early recognition of Arg deficiencies. Furthermore, despite being a downstream product of Arg catabolism and their many functions within the cell, polyamines were not a limiting factor for the growth of ASS1 deficient cells in a low Arg environment as we expected. However, Orn caused large improvements to cellular growth when supplemented in low Arg media. Because Orn was unable to rescue cells in an Arg free environment, we hypothesize that Orn is being shunted through a non-Arg related pathway to help the cell cope with Arg deficiency coupled with ASS1 downregulation. Therefore, Orn presents as a worthy point of exploration moving forward. Understanding the mechanisms by which cells respond to amino acid deprivation and the metabolic pathways essential to the growth of cancers that are deficient in ASS1 could reveal new vulnerabilities used in their treatment.
References
- Faubert, B., Solmonson, A., & DeBerardinis, R. J. (2020). Metabolic reprogramming and cancer progression. Science, 368(6487). https://doi.org/10.1126/science.aaw5473
- Chen, C.-L., Hsu, S.-C., Ann, D. K., Yen, Y., & Kung, H.-J. (2021). Arginine signaling and cancer metabolism. Cancers, 13(14), 3541. https://doi.org/10.3390/cancers13143541
- Cheng, C.-T., Qi, Y., Wang, Y.-C., Chi, K. K., Chung, Y., Ouyang, C., Chen, Y.-R., Oh, M. E., Sheng, X., Tang, Y., Liu, Y.-R., Lin, H. H., Kuo, C.-Y., Schones, D., Vidal, C. M., Chu, J. C.- Y., Wang, H.-J., Chen, Y.-H., Miller, K. M., … Ann, D. K. (2018). Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Communications Biology, 1(1). https://doi.org/10.1038/s42003-018-0178-4
- Delage, B., Fennell, D. A., Nicholson, L., McNeish, I., Lemoine, N. R., Crook, T., & Szlosarek, P. W. (2010). Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. International Journal of Cancer. https://doi.org/10.1002/ijc.25202
- Keshet, R., Szlosarek, P., Carracedo, A., & Erez, A. (2018). Rewiring urea cycle metabolism in cancer to support anabolism. Nature Reviews Cancer, 18(10), 634–645. https://doi.org/10.1038/s41568-018-0054-z
- Butler, M., van der Meer, L. T., & van Leeuwen, F. N. (2021). Amino acid depletion therapies: Starving cancer cells to death. Trends in Endocrinology & Metabolism, 32(6), 367–381. https://doi.org/10.1016/j.tem.2021.03.003
- Long, Y., Tsai, W.-B., Wangpaichitr, M., Tsukamoto, T., Savaraj, N., Feun, L. G., & Kuo, M. T. (2013). Arginine deiminase resistance in melanoma cells is associated with metabolic reprogramming, glucose dependence, and glutamine addiction. Molecular Cancer Therapeutics, 12(11), 2581–2590. https://doi.org/10.1158/1535-7163.mct-13-0302
- Apiz Saab, J. J., Dzierozynski, L. N., Jonker, P. B., AminiTabrizi, R., Shah, H., Menjivar, R. E., Scott, A. J., Nwosu, Z. C., Zhu, Z., Chen, R. N., Oh, M., Sheehan, C., Wahl, D. R., Pasca di Magliano, M., Lyssiotis, C. A., Macleod, K. F., Weber, C. R., & Muir, A. (2023). Pancreatic tumors exhibit myeloid-driven amino acid stress and upregulate arginine biosynthesis. eLife, 12. https://doi.org/10.7554/elife.81289
- Kremer, J. C., Prudner, B. C., Lange, S. E., Bean, G. R., Schultze, M. B., Brashears, C. B., Radyk, M. D., Redlich, N., Tzeng, S.-C., Kami, K., Shelton, L., Li, A., Morgan, Z., Bomalaski, J. S., Tsukamoto, T., McConathy, J., Michel, L. S., Held, J. M., & Van Tine, B. A. (2017). Arginine deprivation inhibits the Warburg effect and upregulates glutamine anaplerosis and serine biosynthesis in ASS1-deficient cancers. Cell Reports, 18(4), 991–1004. https://doi.org/10.1016/j.celrep.2016.12.077
- Lamming, D. W. (2016). Inhibition of the mechanistic target of rapamycin (mtor)–rapamycin and beyond. Cold Spring Harbor Perspectives in Medicine, 6(5). https://doi.org/10.1101/cshperspect.a025924
- Ennis, H. L., & Lubin, M. (1964). Cycloheximide: Aspects of inhibition of protein synthesis in mammalian cells. Science, 146(3650), 1474–1476. https://doi.org/10.1126/science.146.3650.1474
- Masson, G. R. (2019). Towards a model of GCN2 activation. Biochemical Society Transactions, 47(5), 1481–1488. https://doi.org/10.1042/bst20190331
- Averous, J., Lambert-Langlais, S., Mesclon, F., Carraro, V., Parry, L., Jousse, C., Bruhat, A., Maurin, A.-C., Pierre, P., Proud, C. G., & Fafournoux, P. (2016). GCN2 contributes to mtorc1 inhibition by leucine deprivation through an ATF4 independent mechanism. Scientific Reports, 6(1). https://doi.org/10.1038/srep27698
- Shen, S., Kai, B., Ruan, J., Torin Huzil, J., Carpenter, E., & Tuszynski, J. A. (2006). Probabilistic analysis of the frequencies of amino acid pairs within characterized protein sequences. Physica A: Statistical Mechanics and Its Applications, 370(2), 651–662. https://doi.org/10.1016/j.physa.2006.03.004
- Sekhar, Vandana, et al. “ATP13A3 Facilitates Polyamine Transport in Human Pancreatic Cancer Cells.” Scientific Reports, vol. 12, no. 1, 2022, https://doi/org/10.1038/s41598-02207712-4.
- Hamouda, N. N., Van den Haute, C., Vanhoutte, R., Sannerud, R., Azfar, M., Mayer, R., Cortés Calabuig, Á., Swinnen, J. V., Agostinis, P., Baekelandt, V., Annaert, W., Impens, F., Verhelst, H. L., Eggermount, J., Martin, S., & Vangheluwe, P. (2021). ATP13A3 is a major component of the enigmatic mammalian polyamine transport system. Journal of Biological Chemistry, 296, 100182. https://doi.org/10.1074/jbc.ra120.013908
- Sørensen, D. M., Holemans, T., van Veen, S., Martin, S., Arslan, T., Haagendahl, I. W., Holen, H. W., Hamouda, N. N., Eggermount, J., Palmgren, M., & Vangheluwe, P. (2018). Parkinson disease related ATP13A2 evolved early in animal evolution. PLOS ONE, 13(3).
- Sagar, Narashans Alok, et al. “Polyamines: Functions, Metabolism, and Role in Human Disease Management.” Medical Sciences, vol. 9, no. 2, 2021, p. 44, https://doi.org/10.3390/medsci9020044.
- Ginguay, Antonin, et al. “Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways.” Biology, vol. 6, no. 4, 2017, p. 18, https://doi.org/10.3390/biology6010018

