Spencer Fox Eccles School of Medicine
68 Overexpression of GLUL Rescues Cell Growth in Glutamine Depleted Environments
Libby Brooks
Faculty Mentor: Natasha Pavlova (Oncological Sciences, University of Utah)
About 90% of cancer deaths are from the development of secondary tumor growths from a process called metastasis. As cancer cells divide, they deplete surrounding tissue of nutrients, especially the amino acid glutamine. Most cells cannot survive without glutamine. However, even in glutamine depleted environments, tumors have found ways to grow. Tumors are comprised of cancerous cells as well as non-cancerous stromal cells.
Studies have shown stromal cells can synthesize glutamine which they release into the tumor microenvironment. This feeds cancer cells and allows them to grow. Glutamine synthetase enzyme (GLUL) synthesizes glutamine. However, it is unknown if GLUL activity alone can synthesize enough glutamine to support cell growth in glutamine deficient environments.
To study the GLUL activity, cells were genetically modified to overexpress the enzyme and grown in conditions without glutamine. The hypothesis for this project is that GLUL expression will rescue the growth of these cells. GLUL expression may be able to promote growth of cells even in the absence of environmental glutamine availability. The results of this study agree with the hypothesis. This understanding will aid in the development of cancer therapeutics to treat metastatic cancers.
Index terms – glutamine, glutamine synthetase (GLUL), metastasis, tumor microenvironment (TME)
I. IntroductionAbout 90% of cancer deaths are from secondary tumor growths called metastasis [1]. Understanding the tumor microenvironment and cancer cell proliferation is crucial for advancing cancer treatments. Tumors are made of both cancerous cells as well as non- cancerous stromal cells [2]. The relationship between these cells is critical for tumor growth.
The most limiting factor for tumor growth is amino acid availability, especially the amino acid glutamine, not energy [3]. Due to poor vasculature, the tumor microenvironment is quickly depleted of amino acids used to support cell metabolism that would normally be supplied by blood [4]. The most quickly depleted amino acid in the tumor microenvironment is glutamine [5], [6], [7]. Most mammalian cells cannot survive or proliferate without glutamine [8]. Under stressful conditions, healthy, stromal cells, such as cancer-associated-fibroblasts, have been shown to synthesize glutamine [9], [10]. Cancer cells however do not produce glutamine making them metabolically dependent on stromal cells [2].
Cells synthesize non-essential amino acids using enzymes. Glutamate-ammonia ligase (GLUL) is an enzyme that synthesizes glutamine [8]. In the presence of glutamine, GLUL is unstable and is broken down [11]. Thus, GLUL expression increases in the absence of glutamine causing the cell to synthesize it. Although cell growth in the tumor microenvironment is most limited by glutamine, it is unclear if GLUL activity alone is enough to rescue cell growth.
This project tests the hypothesis that the overexpression of GLUL will rescue the growth of cells grown in glutamine deficient culture media.
Methods
A. Cell Culture
All cell lines were cultured at 37° in a 5% CO2 incubator. For cells cultured with glutamine, glutamine free MEM-Alpha media was prepared with 10% FBS, 1% penicillin- streptomycin and 2 mM glutamine. For cells cultured without glutamine, glutamine free MEM- Alpha media was prepared with 10% dialyzed fetal bovine serum (dFBS) and 1% penicillin- streptomycin. CRISPR modified pTURN lines used doxycycline as an inducer and were treated with 1ug/mL concentrations.
Two cells lines were cultured for these experiments. As a healthy cell model, 10T1/2s were used. 10T1/2s are mouse mesenchymal progenitor cells with fibroblast characteristics. These cells are immortalized mouse stem cells. As a cancerous model, the mouse mammary carcinoma line E0771 was used.
B. Genetic Modifications
Retroviral constructs expressing HA-tagged GLUL or empty vector control were provided by the Pavlova Lab. Cells were modified for the overexpression of GLUL (pTURN GLUL HA) to show that GLUL is rescuing cell growth in glutamine deficient environments rather than other processes such as obtaining glutamine through autophagy, as well as a control (pTURN vec). These cell lines are to test the hypothesis that increased levels of GLUL alone is sufficient to allow cells to synthesize enough glutamine to rescue their growth.
C. Glutamine Deprivation
Cell growth was used as an indicator of metabolic activity for cell rescue or death. To study the metabolic reliance of cells on glutamine cells were plated in triplicates in 12 well dishes at a concentration of 30k/well with media containing 2 mM glutamine. After 24 hrs, day one triplicates were counted using Beckman Coulter counter to compare relative growth of other conditions to. The remaining wells were washed with MEM-Alpha without glutamine and treated with two conditions, 2 mM glutamine or 0 mM glutamine. Cells treated with 2mM glutamine conditions were counted on day 3 after plating and 0 mM glutamine conditions were counted on day 4 after plating. The relative growth of cells was determined by dividing the number of cells in each well by the average number of cells from the day one counts.
D. Western Blotting
Western blots were performed to confirm CRISPR modified lines as well as to compare upregulated proteins in different cell treatments. Cells were prepared for western blotting by being plated in 6 cm dishes as concentrations of 300k cells/well. Cells were grown in 2 mM glutamine MEM-Alpha for the first 24 hours. After 24 hours the cells were washed with 0 mM glutamine media and treated with the same treatments as the glutamine deprivation experiments for 24 hours before extracting proteins for western blots.
Proteins were extracted using 1xRIPA buffer, protease inhibitor and phosphatase inhibitor. The total harvested protein concentrations were determined with a BCA assay. Equal amounts of protein were pipetted into wells a 4-12% Bis-Tris gel.
Four proteins were blotted for GLUL, total GCN2, charged GCN2 and vinculin. GLUL is to identify if cells are synthesizing their own glutamine to proliferate in glutamine deficient conditions. GCN2 indicates amino acid deficiency by responding to uncharged tRNA. Total levels of protein and charged levels of protein were both measured to compare relative amounts of charged protein in the cell. Vinculin was used as a loading control.
E. Statistical Analysis
Statistical analysis was done using two tailed t tests on relative cell growth numbers. In glutamine deprivation experiments, 0 mM and 2 mM glutamine conditions were compared for statistically significant differences (p < 0.05) in growth. All experiments were repeated 3-5 times.
Results
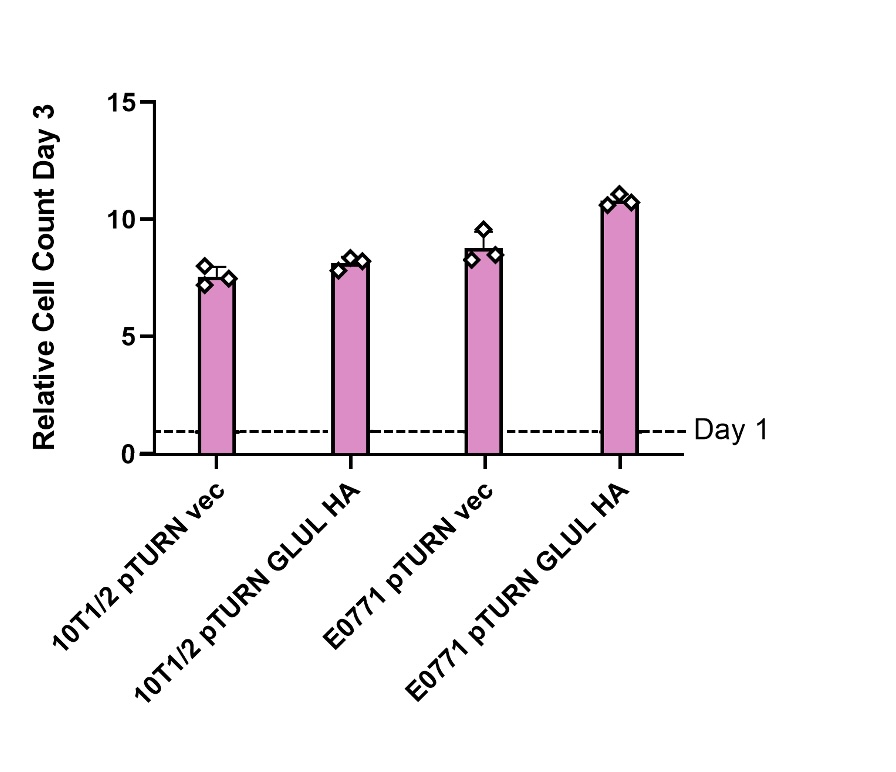
A. Overexpression of GLUL did not affect growth in media containing glutamine
Cells genetically modified to overexpress GLUL (pTURN GLUL HA) and a control (pTURN vec) genotype were grown in media containing 2 mM glutamine (see Figure 1). In both healthy 10T1/2 cells and mouse mammary carcinoma E0771 cells comparing the control and GLUL overexpression there was no statistical difference in growth (see Figure 1). This showed that GLUL does not effect cell growth in the presence of glutamine.
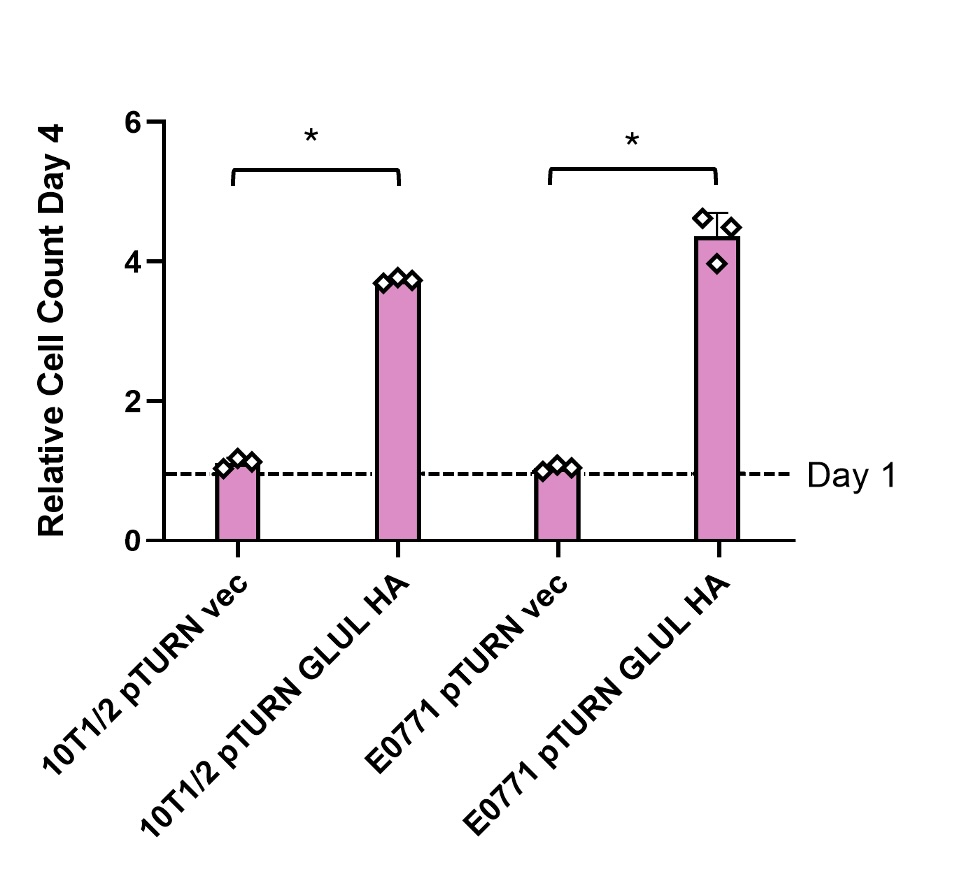
B. Over-expression of GLUL rescued cell growth in glutamine deficient media
Stromal cells and cancer cells were both able to produce enough glutamine to rescue growth with increased expression of GLUL in 0 mM glutamine media (see Figure 2). Non- cancerous 10T1/2 cells that did not overexpress GLUL were still able to grow some without glutamine. Cancerous E0771 cells were not able to grow at all without the GLUL overexpression genetic modification.
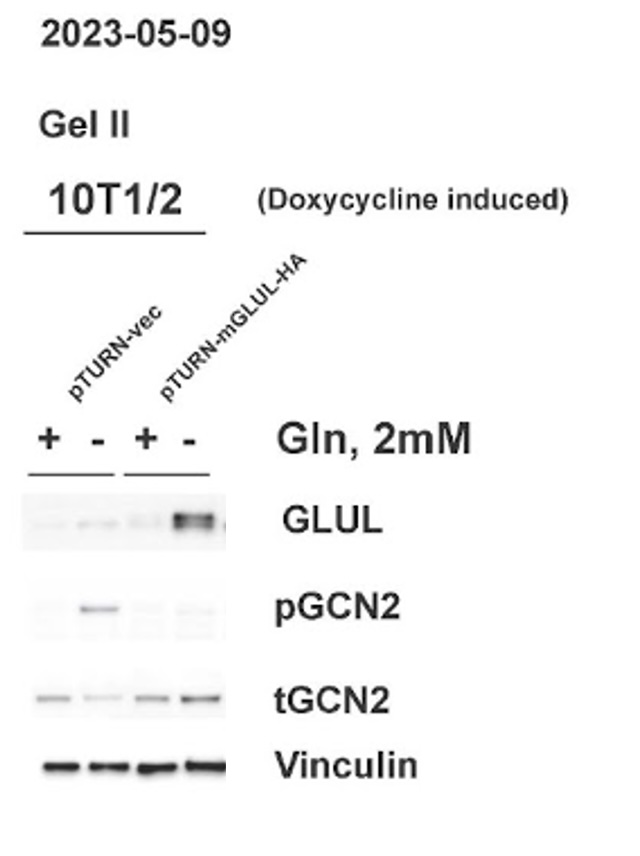
C. GLUL over-expression led to a decrease in uncharged tRNA
Western blot analysis confirmed that cells with the pTURN GLUL HA genotype had an increased expression of GLUL. The western blot also showed the increase of pGCN2 in 0 mM glutamine media observed in the control condition did not happen in cells overexpressing GLUL. The levels of tGCN2 remained similar across all conditions.
Discussion
This project studies the ability of glutamine synthetase (GLUL) to rescue cell growth in glutamine deficient environments. Glutamine is the first depleted amino acid in the tumor microenvironment and thus the most limiting factor for cell growth [8]. It was hypothesized that GLUL activity alone can synthesize enough glutamine for cells to grow in glutamine depleted media. Previous literature has shown that when glutamine is depleted, non-cancerous stromal cells, such as cancer associated fibroblasts (CAFs), begin to synthesize glutamine that is released into the environment [9]. An experiment was designed to test if GLUL expression is enough to rescue cells grown in glutamine deprived conditions similar to the tumor microenvironment.
Non-cancerous stromal cells (10T1/2) and mouse mammary carcinoma cells (E0771) both grew normally in media with 2 mM glutamine (Figure 1). This shows that the expression of GLUL had no measurable effects on the ability of cells to grow when there was environmental glutamine available. Western blots also showed that there was not an increase in GLUL even in cells with the GLUL overexpression genetic modification (Figure 3). This result was expected because there is no need for cells to express GLUL if the environmental glutamine supply is sufficient for cell activities. In this condition GLUL will not be stable in the cell and will be broken down [11]. 10T1/2 and E0771 cells with the pTURN-GLUL-HA modification grew more than the pTURN-vec modification in 0 mM glutamine (Figure 2). This shows that with the overexpression of GLUL, cells are not dependent on environmental glutamine. Western blots showed that in the pTURN-vec cells there was an increase in pGCN2 (Figure 3). pGCN2 is an indicator of uncharged tRNA. This shows that growth in these cells was limited by an amino acid deficiency. This can be explained by the fact that these cells also did not have an increase in GLUL expression. When GLUL expression increased, there was not an increase in uncharged tRNA and cells continued to grow. This provides evidence that it is GLUL activity that is rescuing the growth of these cells.
A limitation of this study and potential area of future work is quantifying the glutamine that cells are secreting. Although these results show that cells are synthesizing and secreting glutamine, how much remains unclear. Quantifying the amount of glutamine these cells are making could provide more insight into the ability of cells to replenish glutamine supplies in the tumor microenvironment.
Showing that GLUL activity can rescue cell growth and that cells expressing GLUL are not dependent on environmental glutamine provides more understanding for the tumor microenvironment. Having a better understanding of how nutrients are supplied to tumors in early growth will help identify cellular processes to target for future cancer therapeutics. This especially relevant in cases of metastatic cancer when new tumors begin to form. Being able to limit the about of glutamine that cancer cells have available to them will inhibit their ability to grow. Knowing that GLUL is an essential part of that pathway will allow future work to develop ways to control the amount of glutamine available in the tumor microenvironment.
References
[1] X. Guan, “Cancer metastases: challenges and opportunities,” Acta Pharm. Sin. B, vol. 5, no. 5, Art. no. 5, Sep. 2015, doi: 10.1016/j.apsb.2015.07.005.
[2] M. Tajan and K. H. Vousden, “The Quid Pro Quo of the Tumor/Stromal Interaction,” Cell Metab., vol. 24, no. 5, Art. no. 5, Nov. 2016, doi: 10.1016/j.cmet.2016.10.017.
[3] L. W. S. Finley, “What is cancer metabolism?,” Cell, vol. 186, no. 8, Art. no. 8, Apr. 2023, doi: 10.1016/j.cell.2023.01.038.
[4] F. F. Diehl et al., “Nucleotide imbalance decouples cell growth from cell proliferation,”
Nat. Cell Biol., vol. 24, no. 8, Art. no. 8, Aug. 2022, doi: 10.1038/s41556-022-00965-1.
[5] M. Pan et al., “Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation,” Nat. Cell Biol., vol. 18, no. 10, Art. no. 10, Oct. 2016, doi: 10.1038/ncb3410.
[6] N. N. Pavlova et al., “Translation in amino-acid-poor environments is limited by tRNAGln charging,” eLife, vol. 9, p. e62307, Dec. 2020, doi: 10.7554/eLife.62307.
[7] N. Pavlova et al., “As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid,” Cell Metab., vol. 27, no. 2, pp. 428-438.e5, Feb. 2018, doi: 10.1016/j.cmet.2017.12.006.
[8] Zhang, N. N. Pavlova, and C. B. Thompson, “Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine,” EMBO J., vol. 36, no. 10, Art. no. 10, May 2017, doi: 10.15252/embj.201696151.
[9] Sahai et al., “A framework for advancing our understanding of cancer-associated fibroblasts,” Nat. Rev. Cancer, vol. 20, no. 3, Art. no. 3, Mar. 2020, doi: 10.1038/s41568- 019-0238-1.
[10] -Y. He et al., “Chronic stress increases metastasis via neutrophil-mediated changes to the microenvironment,” Cancer Cell, vol. 0, no. 0, Feb. 2024, doi: 10.1016/j.ccell.2024.01.013.
[11] Șafak Bayram et al., “Investigating the role of GLUL as a survival factor in cellular adaptation to glutamine depletion via targeted stable isotope resolved metabolomics,” Mol. Biosci., vol. 9, p. 859787, Aug. 2022, doi: 10.3389/fmolb.2022.859787.

