John and Marcia Price College of Engineering
11 Three-Dimensional Construction of Coronary Vasculature Geometries
Aksel Anderson
Faculty Mentor: Rob MacLeod (Bioengineering, University of Utah)
Introduction
Cardiovascular disease is the leading cause of death globally, and one of the most impactful subsets is coronary artery disease (CAD) [1]. CAD develops due to a buildup of plaque in the arterial walls, which narrows the arterial lumen and restricts blood flow. Obstruction(s) in the coronary arteries reduce the supply of oxygen and nutrients to the heart muscle, known as the myocardium [2]. The deficiency in blood flow to the myocardium can lead to what is known as myocardial ischemia (MI). An ischemic myocardium may not function properly, potentially compromising the heart’s mechanical and electrical function.
Furthermore, severe MI can lead to cell death [3]. Examining the heart’s vascular network is one way researchers can study CAD and its cascading effects on the development of MI [4]. However, past studies reconstructing patient-specific vasculature geometries have been limited to the main branches, neglecting the smaller downstream branches [5]. Capturing downstream vasculature would improve our understanding of the negative consequences of reduced blood flow to the myocardium [6]. Our study produced a novel method to obtain subject-specific vasculature geometries that include the downstream vasculature.
Methods
We obtained five explanted porcine hearts and administered the novel contrast agent BriteVu (Scarlet Imaging), which highlights at enhanced resolution the coronaries in computed tomography (CT) images. We created three-dimensional vasculature segmentations from the CT images using the open-source software Seg3D [7]. Furthermore, we developed a Python program to remove discontinuities in the resulting segmentations. Once the final geometries were obtained, we computed three metrics to determine the depth of the captured vasculature: (1) the number of vessel segments, (2) the most frequent vessel radius, and (3) the number of vasculature generations, defined by successive levels of branching in the vascular system.
Results
The extent of vasculature captured via our methodology varied across subjects, as shown in Figure 1. Specifically, the number of vessel segments obtained ranged between 74 and 228 vessel segments, with an average of approximately 169 +/- 63 vessel segments. The most frequent vessel radius ranged between 0.30 –
0.64 mm, with an average of approximately 0.44 +/- 0.15 mm. The number of vasculature generations ranged between 6 – 9 generations, with an average of approximately 8 +/- 1 generations.
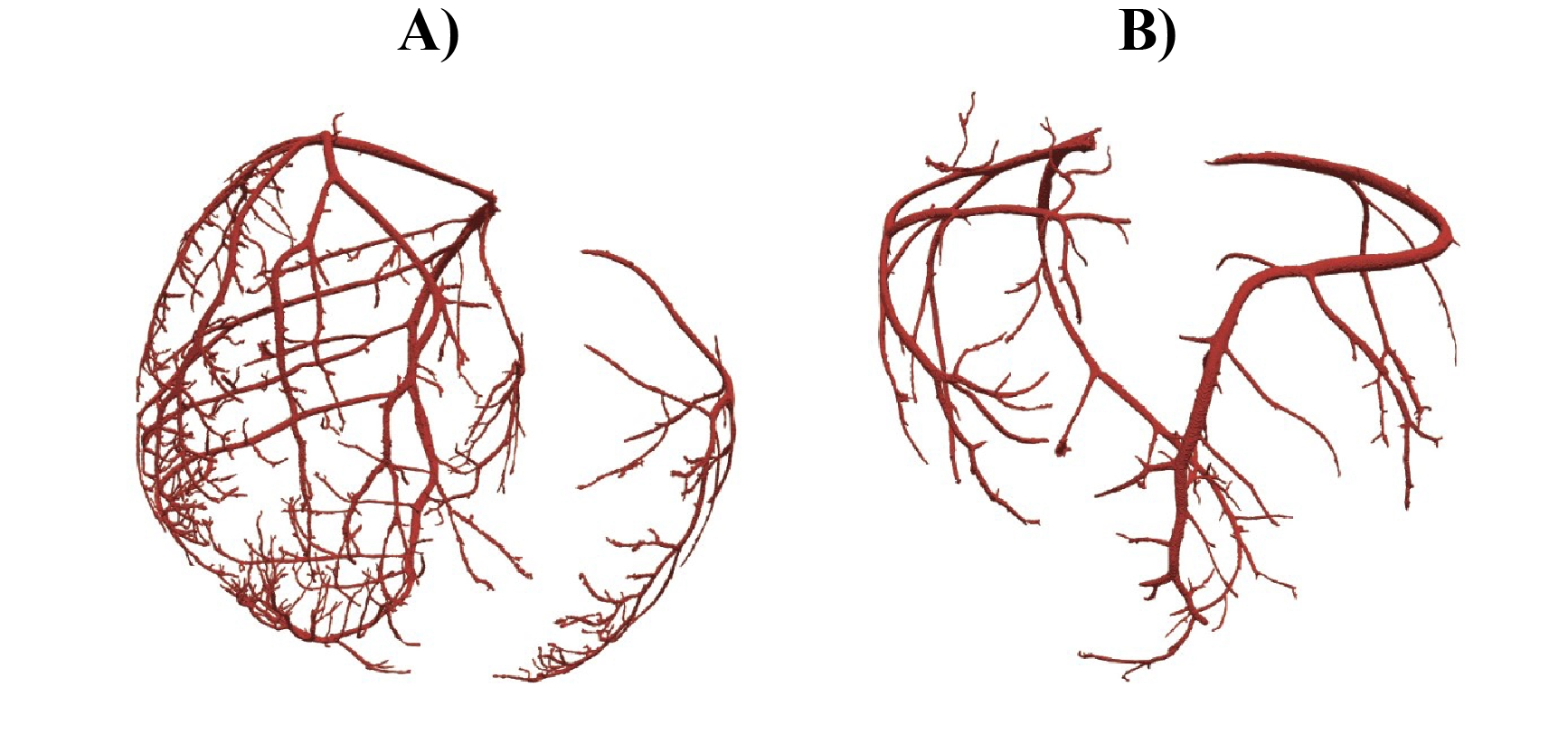
Figure 1. Vasculature Examples. A) Vasculature with 228 vessel segments, a most frequent vessel radius of
0.33 mm, and 9 vasculature generations. B) Vasculature with 74 vessel segments, a most frequent vessel radius of 0.39 mm, and 6 vasculature generations.
Discussion
Our novel methodology provides the ability to study coronary vasculature at unprecedented resolution in a subject-specific manner. The resulting models will provide a substrate to study CAD well beyond the primary branches and hence predict the consequences of narrowing or blockage of a vessel. The average radius of a porcine left coronary artery is approximately 3.5 mm and we were able to successfully capture vessels over 85% smaller than this [8]. However, the number of vessel segments, the most frequent vessel radius, and the number of vasculature generations varied between experiments, showing a potential inconsistency in the downstream vasculature captured in our methodology (Figure 1). Such variability could be attributed to difficulties with the BriteVu injections, resulting in incomplete visualization of the arteries in the CT scan. Our methodology is still limited in identifying the microvasculature, as no known macroscopic imaging approach is capable of capturing vessels at this scale [6]. Our next goal consists of extending these vasculature geometries into the microvasculature level using a rule-based approach to map perfusion beds in the myocardium [9]. We will also overlay the vasculature with distributions of electric potential captured during experiments with acute ischemia. By continuing to refine our methods, we aim to help researchers and clinicians generate complete vascular geometries to enhance the study of CAD.
References
[1] M. A. Khan et al., “Global epidemiology of ischemic heart disease: results from the global burden of disease study,” Cureus, vol. 12, no. 7, p. e9349, doi: 10.7759/cureus.9349.
[2] D. C. Crossman, “The pathophysiology of myocardial ischaemia,” Heart, vol. 90, no. 5, pp. 576–580, May 2004, doi: 10.1136/hrt.2003.029017.
[3] K. Konstantinidis et al., “Mechanisms of cell death in heart disease,” Arterioscler Thromb Vasc Biol., vol. 32, no. 7, pp. 1552–1562, May 2012, doi: 10.1161/ATVBAHA.111.224915.
[4] F. M. A. Nous et al., “Dynamic myocardial perfusion CT for the detection of hemodynamically significant coronary artery disease,” JACC Cardiovasc. Imaging, vol. 15, no. 1, pp. 75–87, Jan. 2022, doi: 10.1016/j.jcmg.2021.07.021.
[5] C. Ada and A. Yong, “Assessment of the coronary microcirculation in the cardiac catheterisation laboratory,” Vessel Plus, vol. 5, no. 57, Dec. 2021, doi: 10.20517/2574-1209.2021.51.
[6] Zhong et al., “Application of patient-specific computational fluid dynamics in coronary and intra- cardiac flow simulations: challenges and opportunities,” Front. Physiol., vol. 9, no. 742, Jun. 2018, doi: 10.3389/fphys.2018.00742.
[7] Seg3D: Volumetric Image Segmentation and Visualization, Scientific Computing and Imaging Institute (SCI) download from: http://www.sci.utah.edu,
[8] Gomez and L. Ballesteros, “Morphologic expression of the left coronary artery in pigs. An approach in relation to human heart,” Rev. Bras. Cir. Cardiovasc., vol. 29, Apr. 2014, doi:10.5935/1678-9741.201400270.
[9] A. Sexton et al., “Rapid model-guided design of organ-scale synthetic vasculature for biomanufacturing,” ArXiv, pp. arXiv:2308.07586v1, Aug. 202

