Transcription and its Regulation in Neurons
Caleb Bevan; Jackson T. Anderson; and Tess Johnson
Chapter under construction. This is the first draft. If you have questions, or want to help in the writing or editing process, please contact hutchins.jim@gmail.com.
Edited by Jim Hutchins on Mar 23 2025
Central Dogma
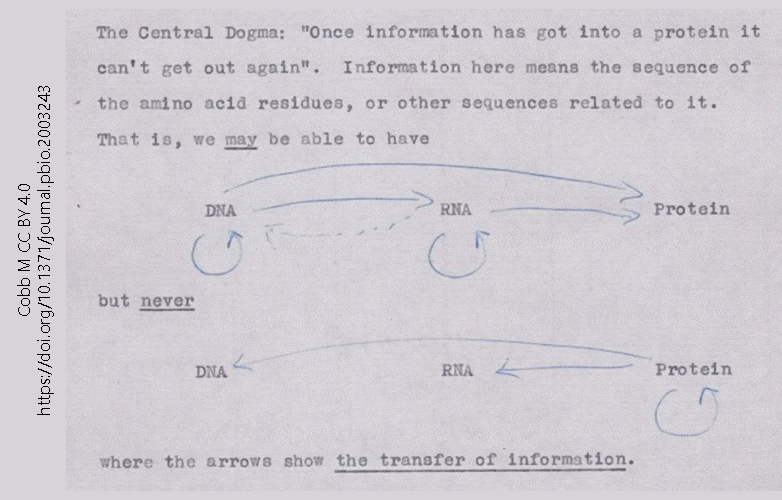 This 1956 note, made by Francis Crick in preparation for a lecture, is the first known statement of what became the Central Dogma of Molecular Biology. The Central Dogma is simple: information flows from DNA to RNA to protein.
This 1956 note, made by Francis Crick in preparation for a lecture, is the first known statement of what became the Central Dogma of Molecular Biology. The Central Dogma is simple: information flows from DNA to RNA to protein.
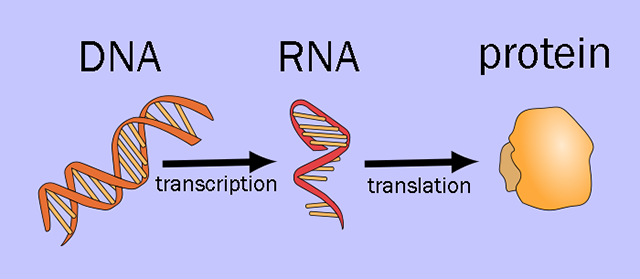
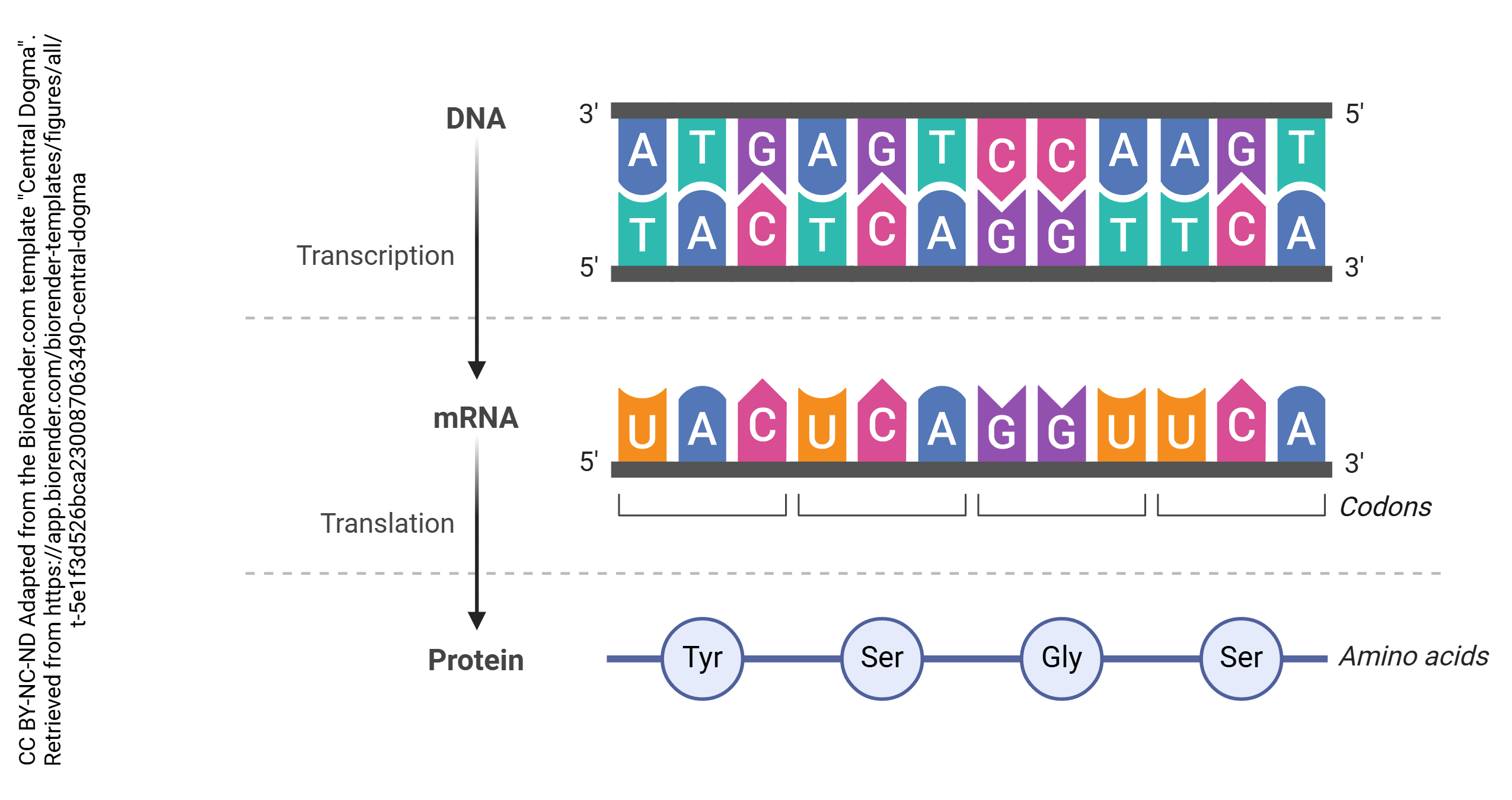
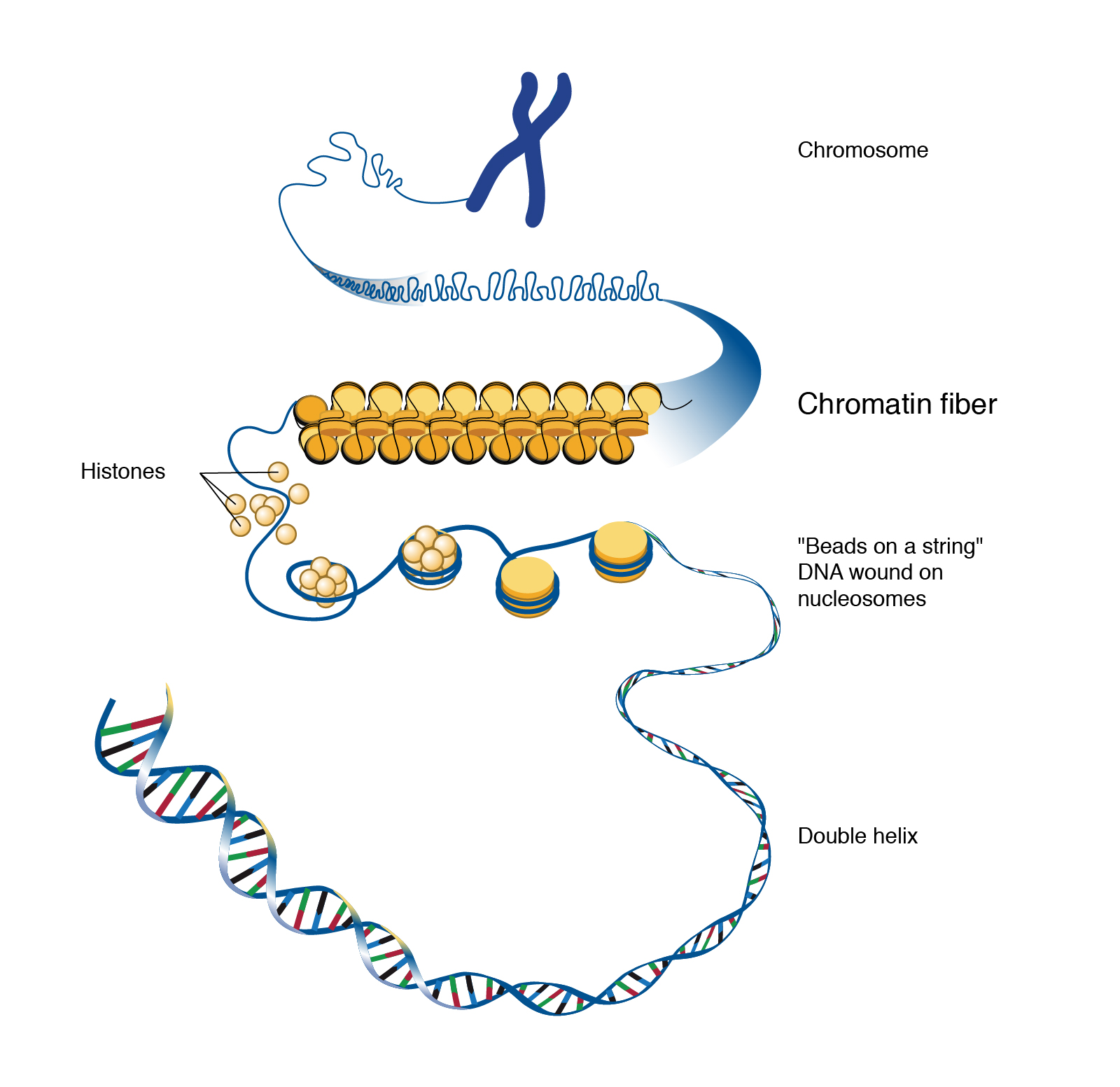
Information in nerve cells is stored in deoxyribonucleic acid (DNA). If unspooled, the DNA in a neuron would be about 3 m long. Because of this, it must be packaged using proteins called histones so it can be packed tightly into each cell.
Genes
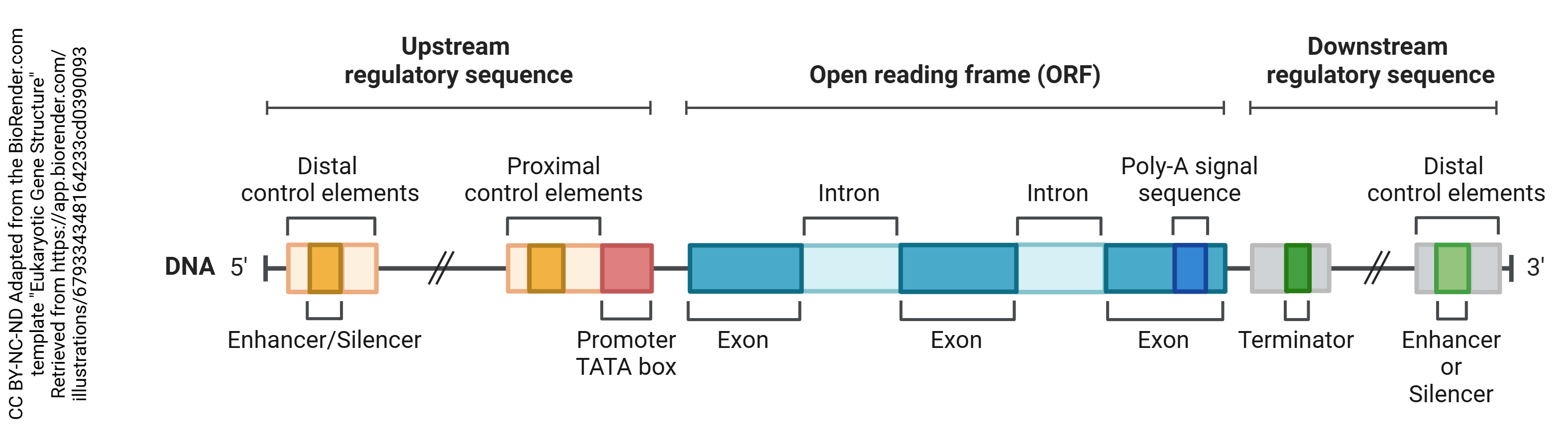
DNA is organized into areas which are used to make proteins or regulatory RNAs. These are collectively called genes. There are about 30,000 genes in the human genome.
Each gene has a region that is read before the actual protein coding region. Because DNA and RNA are always read in the 5-prime to 3-prime direction (5’→3′), which is determined by the chemical structure of nucleic acids, the area before the protein coding region is called the 5′ untranslated region (5′-UTR) or the upstream regulatory sequence and the area after the protein coding region is called the 3′-UTR or downstream regulatory sequence. The gene itself consists of regions that will be made into protein (exons) and regions that are not used to make protein (introns).
Regulation of Transcription
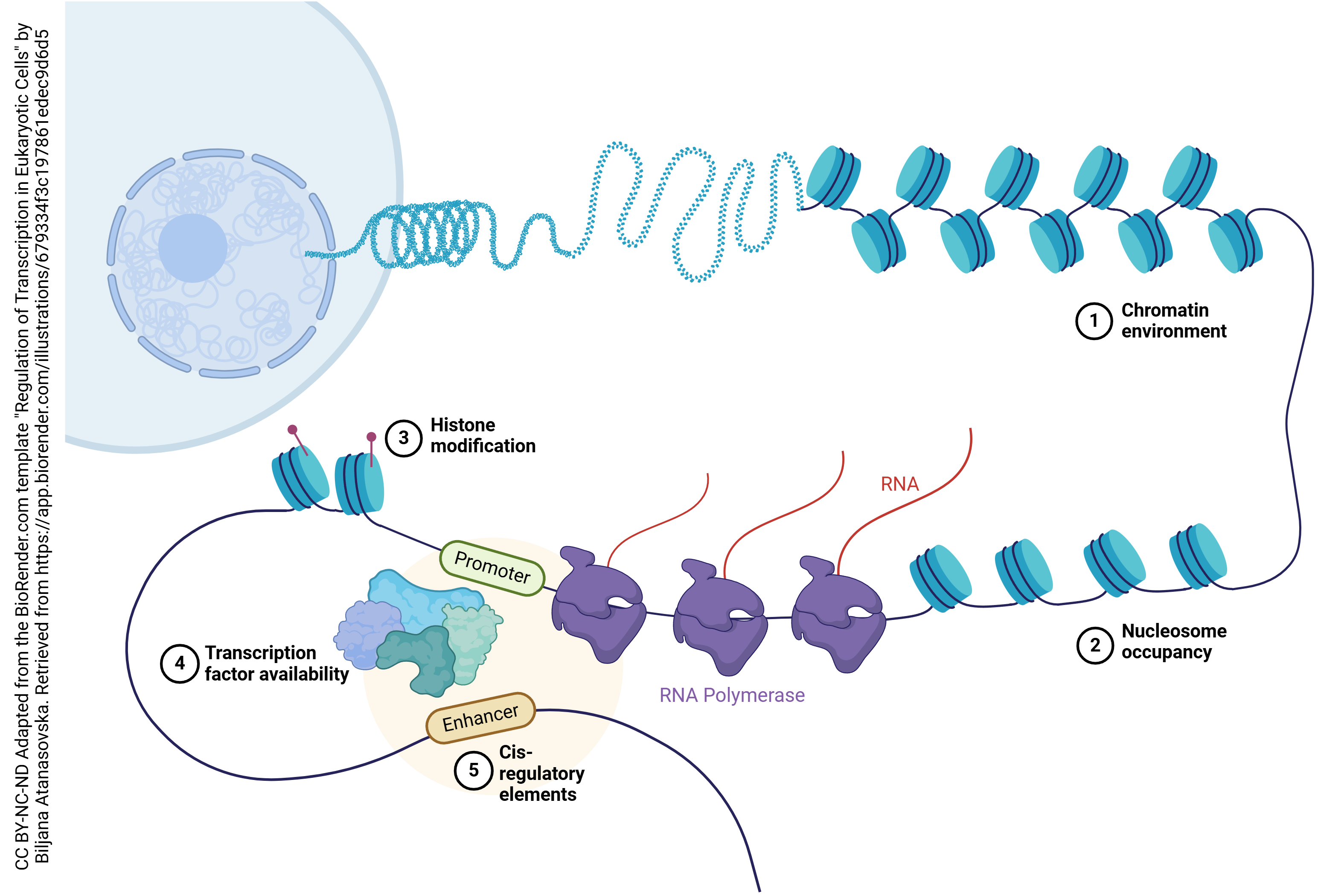
While each neuron has 30,000 genes to choose from, each neuron will choose only a small subset of those genes to be expressed — turned into protein. Genes which are specific to being a liver cell or a kidney cell, for example, are permanently turned off in neurons. Also, most neurons turn off the genes needed to support cell division. To permanently turn off a gene, it is wrapped around histones and left in a compact form which does not permit access to the RNA polymerase.
At an even finer level, neurons will only make receptors (for example) when those receptors are needed to replace those which have “aged out” or are otherwise damaged or lost. This is accomplished through transcriptional regulation. A complex loop is formed which binds to transcription factors, complex assemblies of RNA or protein molecules which help the RNA polymerase find its target.
Copying of DNA into RNA
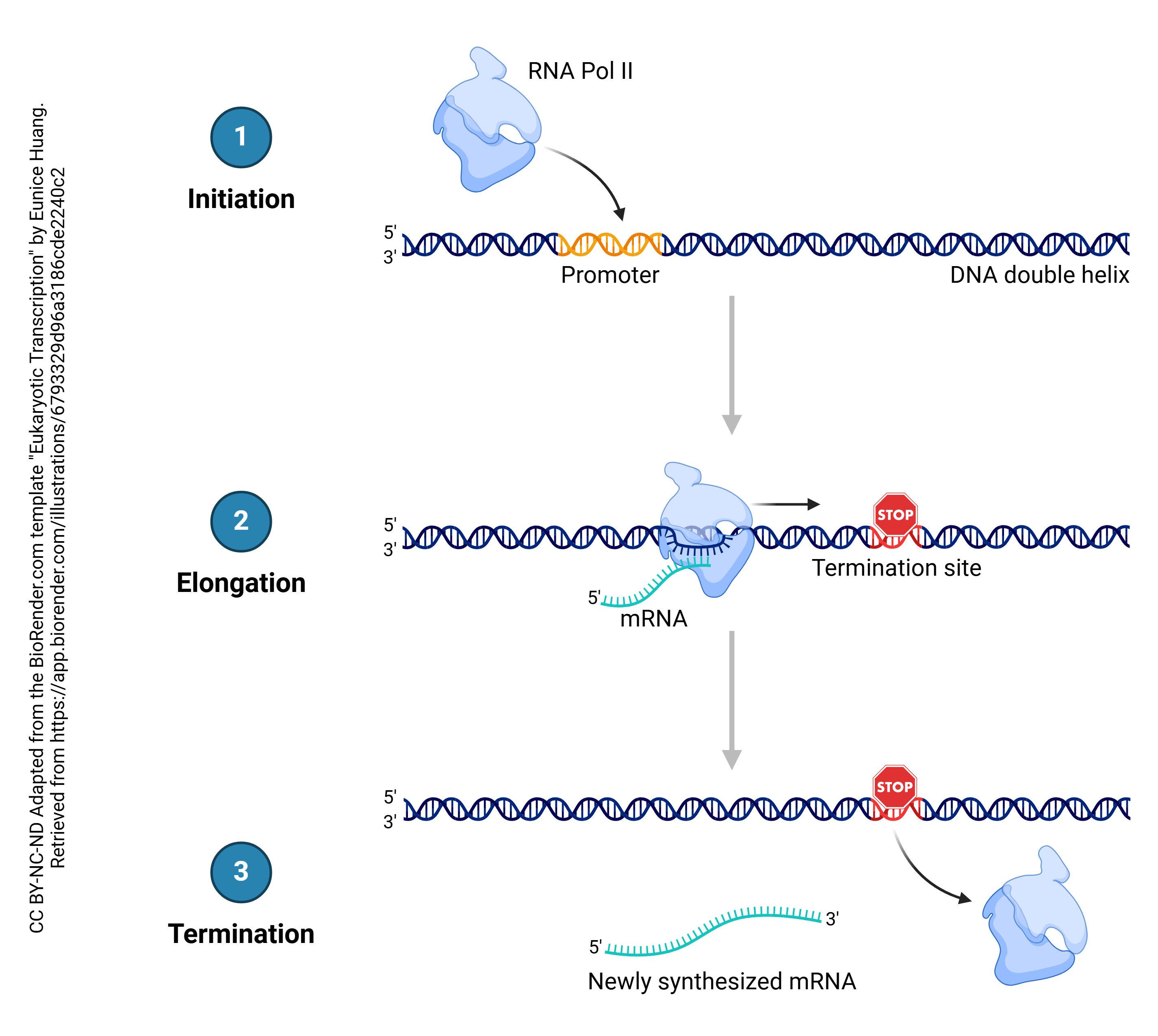
In the process known as transcription, DNA is copied into RNA. In general, DNA is double stranded (i.e. a double helix) while RNA is single-stranded. The enzyme which accomplishes this is RNA polymerase. Bases in the RNA, as it is being made, are matched up to the corresponding bases in DNA, and thymine residues in DNA are replaced by uracil residues in RNA. In that way, each adenine in DNA is matched up to a uracil in RNA; each cytosine is matched up to a guanine; each guanine is matched up to a cytosine; and each thymine in DNA is matched up to an adenine in RNA. This continues until the RNA polymerase encounters the termination site on the DNA double helix.
Splicing
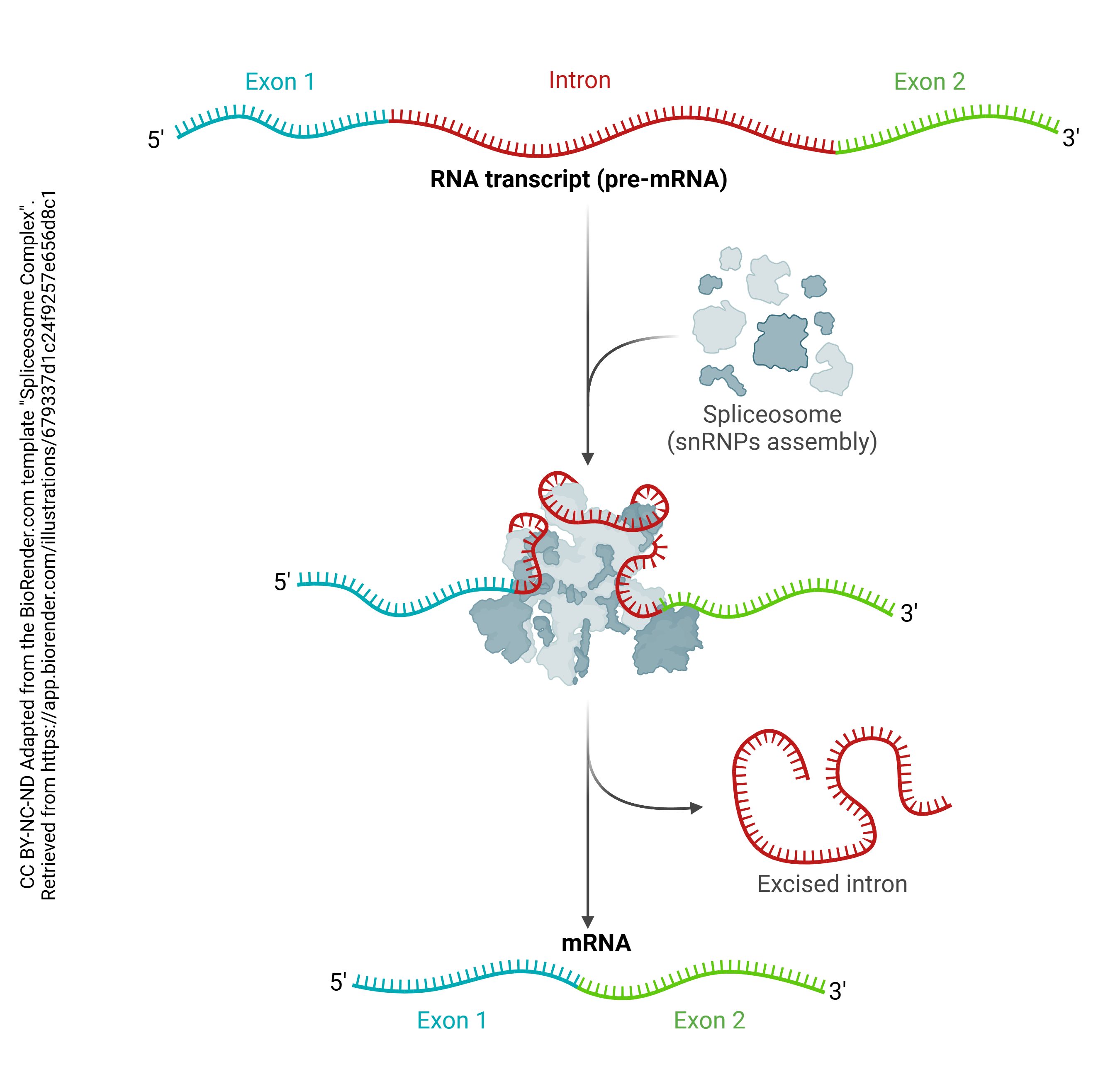
Then after the pre-mRNA (primary transcript) is formed, an organelle called a spliceosome cuts out the RNA which is not part of the protein code (introns) and stitches together the RNA which does code for proteins (exons). A loop of RNA called a lariat is formed, and clipped out. The expressed (i.e. protein coding) regions are tied together to form a string of exons, called messenger RNA (mRNA). It is this mRNA which codes for the protein, as explained in the next chapter. Each mRNA ends in a poly(A) tail: a string of 200 or so adenine residues that will help stabilize the mRNA and ensure that it is translated.
Epigenetics
Permanent chemical alterations in DNA can also change whether genes are more or less likely to be expressed. Because this process is outside of “normal” genetics, it is called epigenetics.
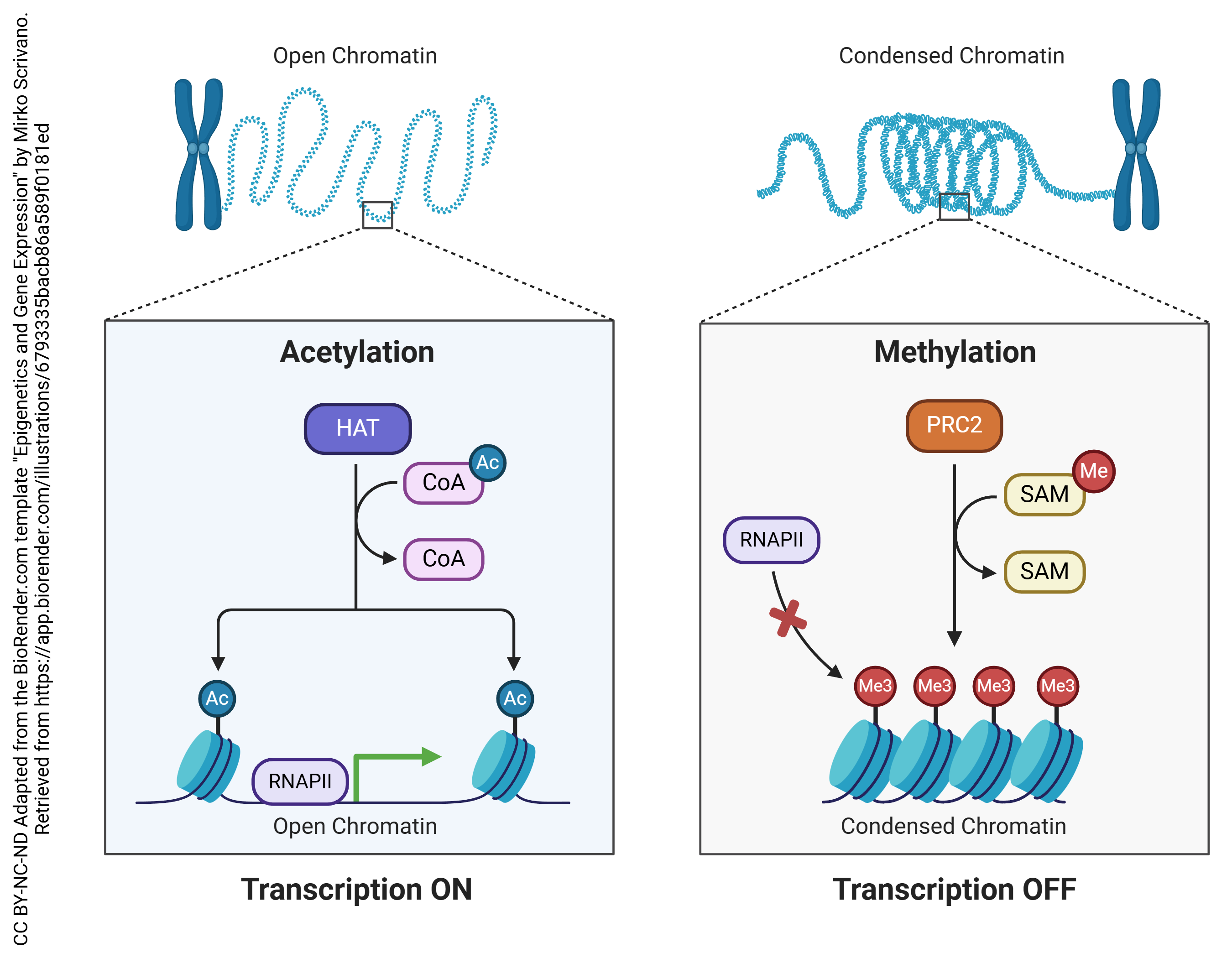
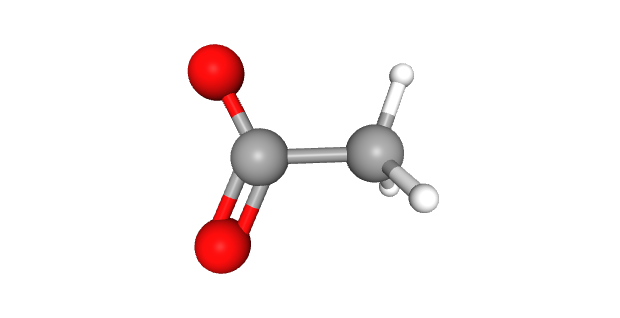
Two of the most common epigenetic modifications of DNA are acetylation and methylation. In acetylation, an acetyl (—CH3COOH) group is added to the DNA, which causes the negatively-charged backbones to repel each other and opens up the configuration of the DNA, making it more accessible to RNA polymerase and therefore more likely to be transcribed.
In methylation, a methyl (—CH3) group is added to cytosine residues on the DNA. This helps compact the DNA structure, making it inaccessible to RNA polymerase and therefore less likely to be transcribed.
Media Attributions
- Central dogma © Francis Crick & Matthew Cobb is licensed under a CC BY (Attribution) license
- Central dogma © Squidonius adapted by Jim Hutchins is licensed under a CC BY-SA (Attribution ShareAlike) license
- Central Dogma © BioRender adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Chromatin © Darryl Leja, NHGRI is licensed under a Public Domain license
- Eukaryotic Gene Structure © Catherine C adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Regulation of Transcription in Eukaryotic Cells © Biljana Atanasovska adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Eukaryotic Transcription © Eunice Huang adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Spliceosome Complex © BioRender adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Epigenetics and Gene Expression © Mirko Scrivano adapted by Jim Hutchins is licensed under a CC BY-NC-ND (Attribution NonCommercial NoDerivatives) license
- Acetate Conformer © PubChem adapted by Jim Hutchins is licensed under a Public Domain license
- Methyl © PubChem adapted by Jim Hutchins is licensed under a Public Domain license

