Epigenetic Contributions to Autism
Adam Evans and Jim Hutchins
Chapter under construction. This is the first draft. If you have questions, or want to help in the writing or editing process, please contact hutchins.jim@gmail.com.
Epigenetics section of Molecular Biology of Neurons chapter in Intro Neuro
Autism spectrum disorder (ASD) is a complex neurodevelopmental condition characterized by differences in social communication, sensory processing, and broader patterns of behavior. While early research emphasized rare genetic mutations as primary contributors to ASD, it has become increasingly clear that epigenetic regulation plays a central role in shaping the presence and phenotypic expression of ASD. In controlling how genes are expressed, epigenetic factors are central to lifelong brain development and synaptic plasticity (see chapter Translation in Neurons). However, epigenetics are especially impactful during early brain development, a critical window for formation of more prevailing patterns of circuitry.
The developing brain undergoes dynamic changes in gene expression regulated by processes such as DNA methylation, histone modification, and non-coding RNAs. These mechanisms act as molecular fine-tuners, enabling precise spatial and temporal control of gene activity. As you may have realized by now, there is no such thing as a “one-size-fits-all” brain. However, there are some specific patterns of disruption to these regulatory layers—whether due to mutation, environmental influence, or stochastic dysregulation—that are associated with the differences in synaptic development and functional connectivity implicated in ASD.
The Autistic Synapse
(Ideally I will phrase this heading more awesome and less stupid)
Keep statistically oriented but emphasize wide phenotypic variation in individuals w/ ASD
[Reference Grijalva & Hutchins chapters]
Mechanisms of Epigenetic Control
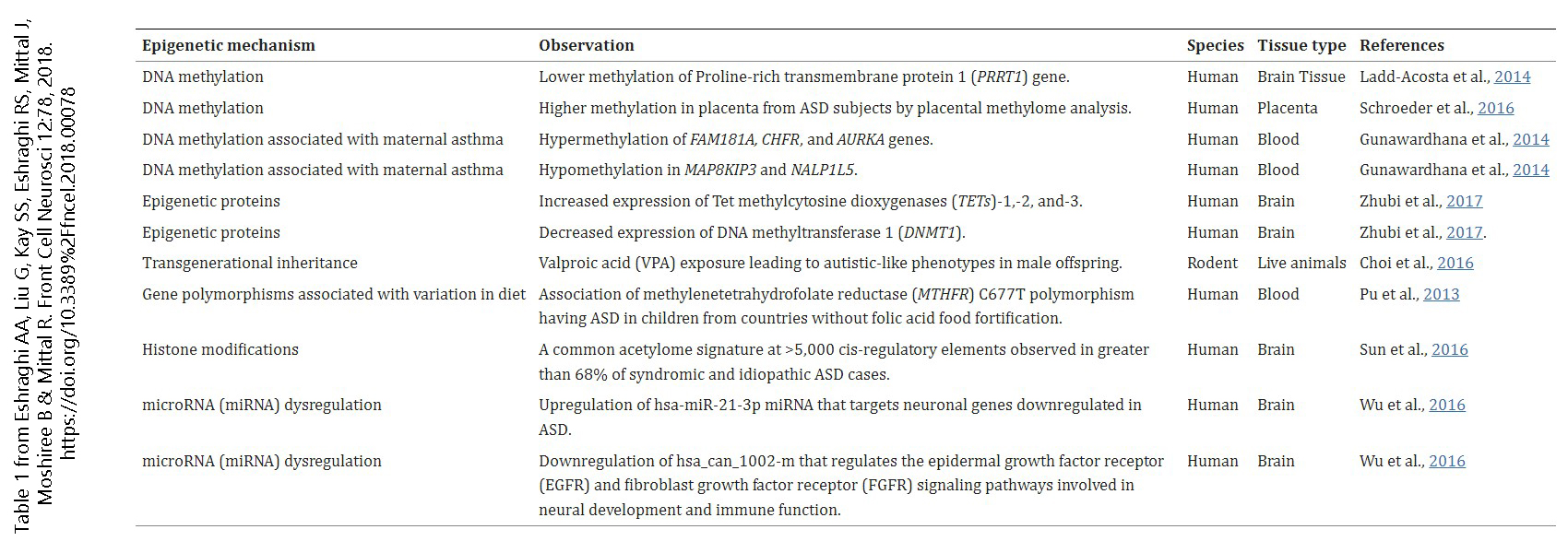 Table from Eshraghi et al.: https://www.frontiersin.org/journals/cellular-neuroscience/articles/10.3389/fncel.2018.00078/full
Table from Eshraghi et al.: https://www.frontiersin.org/journals/cellular-neuroscience/articles/10.3389/fncel.2018.00078/full
DNA methylation is the most widely studied mechanism of ASD’s epigenetic contributing factors, and is popularly considered to be the best understood. [hyper & hypo, include relevant statistics]
Histone acetylation
Histone methylation
Non-coding RNAs https://uen.pressbooks.pub/expertneuro/chapter/translation-in-neuronal-cells/
| Mechanism | Primary Function | Neural Outcome |
|---|---|---|
| DNA Hypermethylation | Directly silences gene expression by adding methyl groups to DNA | Under expression of genes linked to cell cycle control, neural development, |
| DNA Hypomethylation | Lack of DNA methylation compared to general population, leaving “atypical” genes active | Overexpression of genes involved in synaptic signaling, altering neuronal communication; overexpression of some developmental regulators |
| Histone Acetylation | Loosens chromatin to promote transcription | Encourages expression of genes involving ion channel regulation, synaptic function, and neuronal excitability |
| Histone Methylation | Coils chromatin very tightly to make certain genes mechanically inaccessible | Represses genes involved in immune signaling (incl. microglial processes) and HDAC activity |
| Non-Coding RNAs | Post-transcriptional gene regulation | Fine-tunes expression of synaptic and developmental genes |
Critical Periods
- In utero
- Early childhood
- Heightened influences of epigenetic factors during critical periods due to formation of lasting

